
Maribius salinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae;
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
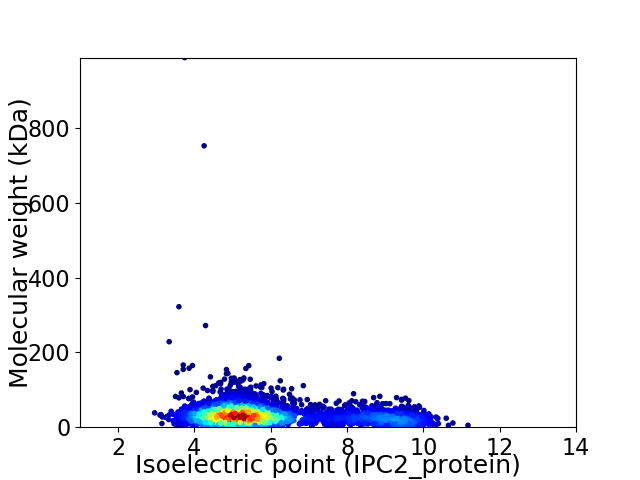
Virtual 2D-PAGE plot for 3413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
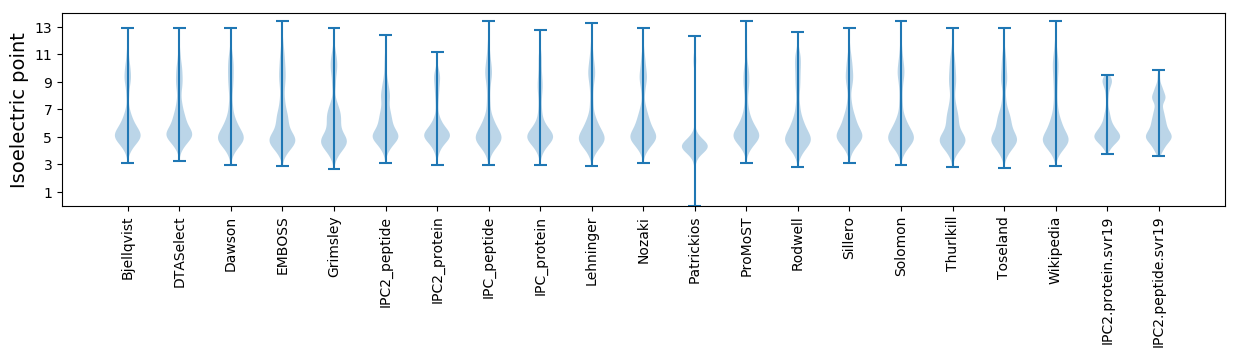
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6AW90|A0A1M6AW90_9RHOB Phosphate transport system permease protein PstA OS=Maribius salinus OX=313368 GN=SAMN04488012_101277 PE=3 SV=1
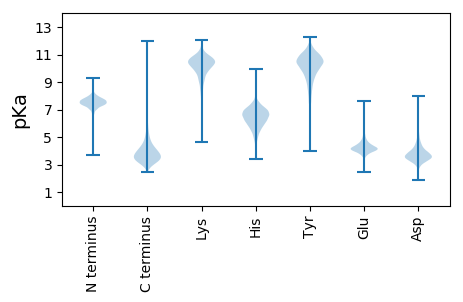
MM1 pKa = 7.49RR2 pKa = 11.84HH3 pKa = 6.09ILLASAATMALGSGMAAAQEE23 pKa = 4.2LVFPVGEE30 pKa = 4.82GAFSWDD36 pKa = 3.34SYY38 pKa = 11.81EE39 pKa = 4.85EE40 pKa = 3.76FAANNDD46 pKa = 3.73YY47 pKa = 11.13SGEE50 pKa = 4.24TIDD53 pKa = 5.87IAGPWTGADD62 pKa = 3.4AEE64 pKa = 4.63LMNSVAAYY72 pKa = 9.27FAEE75 pKa = 4.37ATGATVNYY83 pKa = 9.53SGSDD87 pKa = 3.17SFEE90 pKa = 3.74QDD92 pKa = 2.53IVISTEE98 pKa = 3.9ANSAPNLAAFPQPGLVADD116 pKa = 4.6LASRR120 pKa = 11.84GSITPLDD127 pKa = 3.92EE128 pKa = 3.96EE129 pKa = 4.45TASFIAEE136 pKa = 4.01NYY138 pKa = 9.35SAGEE142 pKa = 4.11SWVDD146 pKa = 3.24LATFEE151 pKa = 5.03GPDD154 pKa = 4.01GEE156 pKa = 4.52SAMYY160 pKa = 10.18AFPYY164 pKa = 10.04KK165 pKa = 9.83IDD167 pKa = 3.59VKK169 pKa = 10.88SLVWYY174 pKa = 10.32SPEE177 pKa = 3.72QFEE180 pKa = 4.29EE181 pKa = 3.94AGYY184 pKa = 9.41EE185 pKa = 3.82IPEE188 pKa = 4.27TYY190 pKa = 10.47EE191 pKa = 3.66EE192 pKa = 4.37LKK194 pKa = 11.01EE195 pKa = 3.88LTAQIAEE202 pKa = 4.54DD203 pKa = 5.67GVTPWCIGLGSGAATGWPATDD224 pKa = 3.1WVEE227 pKa = 5.75DD228 pKa = 3.29IMLRR232 pKa = 11.84TATPEE237 pKa = 3.91QYY239 pKa = 11.2DD240 pKa = 3.36NWVTNDD246 pKa = 3.72MPFDD250 pKa = 4.51DD251 pKa = 4.86PVVVAAIEE259 pKa = 4.28EE260 pKa = 4.62YY261 pKa = 9.86GTFLQEE267 pKa = 4.36GYY269 pKa = 11.18VNGGRR274 pKa = 11.84EE275 pKa = 3.93AAATTDD281 pKa = 4.18FRR283 pKa = 11.84DD284 pKa = 3.76SPSGLFQFPPEE295 pKa = 4.47CYY297 pKa = 8.14MHH299 pKa = 6.63KK300 pKa = 10.01QATFIPTFFPEE311 pKa = 4.4GTVVGEE317 pKa = 4.55DD318 pKa = 2.7VDD320 pKa = 5.26FFYY323 pKa = 11.09FPAPEE328 pKa = 4.84GGEE331 pKa = 4.26TPVLGGGTMFAITEE345 pKa = 4.11DD346 pKa = 3.91SEE348 pKa = 4.45TARR351 pKa = 11.84GFIDD355 pKa = 4.16FLKK358 pKa = 10.23TPIAHH363 pKa = 6.97EE364 pKa = 3.56IWAAQGGLLSPLTSINPEE382 pKa = 3.99VFPSDD387 pKa = 3.33PARR390 pKa = 11.84ATNDD394 pKa = 2.82ILLNATTFRR403 pKa = 11.84FDD405 pKa = 4.8GSDD408 pKa = 3.54LMPGEE413 pKa = 4.38IGAGAFWTQMVEE425 pKa = 4.38FTTGTQDD432 pKa = 3.95AEE434 pKa = 4.3ATATAIQEE442 pKa = 3.94RR443 pKa = 11.84WDD445 pKa = 3.54NLKK448 pKa = 10.6NN449 pKa = 3.5
MM1 pKa = 7.49RR2 pKa = 11.84HH3 pKa = 6.09ILLASAATMALGSGMAAAQEE23 pKa = 4.2LVFPVGEE30 pKa = 4.82GAFSWDD36 pKa = 3.34SYY38 pKa = 11.81EE39 pKa = 4.85EE40 pKa = 3.76FAANNDD46 pKa = 3.73YY47 pKa = 11.13SGEE50 pKa = 4.24TIDD53 pKa = 5.87IAGPWTGADD62 pKa = 3.4AEE64 pKa = 4.63LMNSVAAYY72 pKa = 9.27FAEE75 pKa = 4.37ATGATVNYY83 pKa = 9.53SGSDD87 pKa = 3.17SFEE90 pKa = 3.74QDD92 pKa = 2.53IVISTEE98 pKa = 3.9ANSAPNLAAFPQPGLVADD116 pKa = 4.6LASRR120 pKa = 11.84GSITPLDD127 pKa = 3.92EE128 pKa = 3.96EE129 pKa = 4.45TASFIAEE136 pKa = 4.01NYY138 pKa = 9.35SAGEE142 pKa = 4.11SWVDD146 pKa = 3.24LATFEE151 pKa = 5.03GPDD154 pKa = 4.01GEE156 pKa = 4.52SAMYY160 pKa = 10.18AFPYY164 pKa = 10.04KK165 pKa = 9.83IDD167 pKa = 3.59VKK169 pKa = 10.88SLVWYY174 pKa = 10.32SPEE177 pKa = 3.72QFEE180 pKa = 4.29EE181 pKa = 3.94AGYY184 pKa = 9.41EE185 pKa = 3.82IPEE188 pKa = 4.27TYY190 pKa = 10.47EE191 pKa = 3.66EE192 pKa = 4.37LKK194 pKa = 11.01EE195 pKa = 3.88LTAQIAEE202 pKa = 4.54DD203 pKa = 5.67GVTPWCIGLGSGAATGWPATDD224 pKa = 3.1WVEE227 pKa = 5.75DD228 pKa = 3.29IMLRR232 pKa = 11.84TATPEE237 pKa = 3.91QYY239 pKa = 11.2DD240 pKa = 3.36NWVTNDD246 pKa = 3.72MPFDD250 pKa = 4.51DD251 pKa = 4.86PVVVAAIEE259 pKa = 4.28EE260 pKa = 4.62YY261 pKa = 9.86GTFLQEE267 pKa = 4.36GYY269 pKa = 11.18VNGGRR274 pKa = 11.84EE275 pKa = 3.93AAATTDD281 pKa = 4.18FRR283 pKa = 11.84DD284 pKa = 3.76SPSGLFQFPPEE295 pKa = 4.47CYY297 pKa = 8.14MHH299 pKa = 6.63KK300 pKa = 10.01QATFIPTFFPEE311 pKa = 4.4GTVVGEE317 pKa = 4.55DD318 pKa = 2.7VDD320 pKa = 5.26FFYY323 pKa = 11.09FPAPEE328 pKa = 4.84GGEE331 pKa = 4.26TPVLGGGTMFAITEE345 pKa = 4.11DD346 pKa = 3.91SEE348 pKa = 4.45TARR351 pKa = 11.84GFIDD355 pKa = 4.16FLKK358 pKa = 10.23TPIAHH363 pKa = 6.97EE364 pKa = 3.56IWAAQGGLLSPLTSINPEE382 pKa = 3.99VFPSDD387 pKa = 3.33PARR390 pKa = 11.84ATNDD394 pKa = 2.82ILLNATTFRR403 pKa = 11.84FDD405 pKa = 4.8GSDD408 pKa = 3.54LMPGEE413 pKa = 4.38IGAGAFWTQMVEE425 pKa = 4.38FTTGTQDD432 pKa = 3.95AEE434 pKa = 4.3ATATAIQEE442 pKa = 3.94RR443 pKa = 11.84WDD445 pKa = 3.54NLKK448 pKa = 10.6NN449 pKa = 3.5
Molecular weight: 48.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6GTN6|A0A1M6GTN6_9RHOB TVP38/TMEM64 family membrane protein OS=Maribius salinus OX=313368 GN=SAMN04488012_10584 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.91SLSAA44 pKa = 3.86
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074527 |
39 |
9857 |
314.8 |
33.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.856 ± 0.065 | 0.83 ± 0.018 |
6.868 ± 0.058 | 5.623 ± 0.04 |
3.575 ± 0.028 | 9.194 ± 0.071 |
1.969 ± 0.026 | 4.821 ± 0.033 |
2.574 ± 0.036 | 9.994 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.678 ± 0.032 | 2.327 ± 0.025 |
5.248 ± 0.05 | 2.995 ± 0.025 |
7.161 ± 0.061 | 4.911 ± 0.04 |
5.532 ± 0.051 | 7.373 ± 0.04 |
1.415 ± 0.02 | 2.058 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
