
Marinobacter phage PS3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
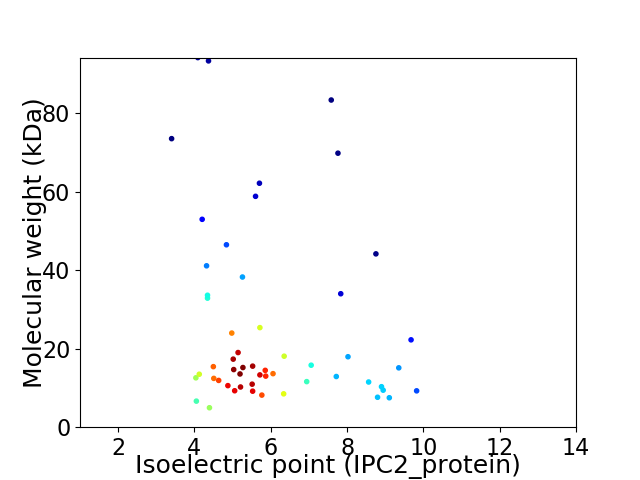
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D1GMR0|A0A2D1GMR0_9CAUD Uncharacterized protein OS=Marinobacter phage PS3 OX=2041341 PE=4 SV=1
MM1 pKa = 7.75ASILGKK7 pKa = 10.64LGIQEE12 pKa = 4.22GDD14 pKa = 3.58SYY16 pKa = 11.16TAAAGSNRR24 pKa = 11.84AVVVAGTGEE33 pKa = 4.22SAFSSRR39 pKa = 11.84EE40 pKa = 3.51IVGITFGGVAMTRR53 pKa = 11.84AAYY56 pKa = 9.13TGDD59 pKa = 3.4TEE61 pKa = 4.34TAKK64 pKa = 11.02GGVYY68 pKa = 10.35YY69 pKa = 9.99ILEE72 pKa = 4.24SEE74 pKa = 4.38IPAGSSVIAVDD85 pKa = 3.06WDD87 pKa = 4.15GQVHH91 pKa = 6.65DD92 pKa = 5.02AFTGAVYY99 pKa = 9.74TLGGIDD105 pKa = 3.87QVTPVDD111 pKa = 3.93NTNSAIDD118 pKa = 3.52VGQFNLDD125 pKa = 3.15ITFTGVDD132 pKa = 3.48GGVLIVGAGSNSPADD147 pKa = 3.83TLNQSSTISATTTTVTVDD165 pKa = 3.32HH166 pKa = 6.41AQITAGHH173 pKa = 6.52EE174 pKa = 4.18GSAISGVLSAAGSEE188 pKa = 4.43TLGLRR193 pKa = 11.84YY194 pKa = 9.81SSEE197 pKa = 4.03TDD199 pKa = 3.25PVGALAVFNSSGAGPSGPSLDD220 pKa = 4.17TVPSTAYY227 pKa = 9.03PGQSRR232 pKa = 11.84TVSGSGFGATQGTGGLTIGGVSQTITSWSDD262 pKa = 2.83TSITFTAVLGANSYY276 pKa = 11.28GSGKK280 pKa = 8.66TIEE283 pKa = 4.29VTTDD287 pKa = 3.74DD288 pKa = 5.42DD289 pKa = 4.17DD290 pKa = 4.28TDD292 pKa = 3.67SGTIEE297 pKa = 4.7LVADD301 pKa = 3.6TAGGFGVVTMSSPNTADD318 pKa = 3.44EE319 pKa = 4.36SSVAYY324 pKa = 9.88GATPTVVTGDD334 pKa = 3.34QFEE337 pKa = 4.57WEE339 pKa = 4.63DD340 pKa = 4.7FNDD343 pKa = 4.18LGNLAIGSDD352 pKa = 3.73AFVSSVDD359 pKa = 3.39TEE361 pKa = 4.34GTFRR365 pKa = 11.84GRR367 pKa = 11.84FWDD370 pKa = 4.15ASDD373 pKa = 3.99GTWGSVNTFTASATDD388 pKa = 3.5EE389 pKa = 4.21TAPTISSASVPTAGNNIAVQMDD411 pKa = 3.14EE412 pKa = 4.14SMQVGAGGTGGWTISLAGVSVSSASVDD439 pKa = 3.48GSDD442 pKa = 3.39DD443 pKa = 3.65TVVNLTPSRR452 pKa = 11.84TLTDD456 pKa = 3.63EE457 pKa = 4.13DD458 pKa = 4.4TLAIGYY464 pKa = 6.09TQPGDD469 pKa = 3.73GFQDD473 pKa = 3.73QADD476 pKa = 4.38TPNDD480 pKa = 3.51LATLSGQAVTNNSAQEE496 pKa = 4.16PPDD499 pKa = 3.12VTGPVTEE506 pKa = 4.59SVDD509 pKa = 3.53VPTGGTYY516 pKa = 10.4AIGDD520 pKa = 3.94DD521 pKa = 5.22LSFTVNWDD529 pKa = 3.23EE530 pKa = 4.12AVTVTGTPALNLDD543 pKa = 3.09IGGTSRR549 pKa = 11.84QADD552 pKa = 3.93YY553 pKa = 11.45ASGSGSSALVFTYY566 pKa = 9.35TVQSGDD572 pKa = 3.48EE573 pKa = 4.41DD574 pKa = 4.8DD575 pKa = 4.93NGIAVSSLTLEE586 pKa = 4.55GGTLQDD592 pKa = 3.58ASSNNATLTLNSVGDD607 pKa = 3.8TSGVLVDD614 pKa = 4.9GVAPVISINALTTTDD629 pKa = 3.46TTPVVTGSAEE639 pKa = 4.1DD640 pKa = 3.89ADD642 pKa = 4.57SLVLDD647 pKa = 4.66LEE649 pKa = 4.87GVDD652 pKa = 3.55ITHH655 pKa = 6.51SSSYY659 pKa = 10.9NITPSAGSWSQQLPEE674 pKa = 4.37LALGEE679 pKa = 4.2YY680 pKa = 10.79LMTLTGVDD688 pKa = 3.17SAGNEE693 pKa = 4.34TVVTAALNVVEE704 pKa = 5.54SVATSPRR711 pKa = 11.84GLFQPLFRR719 pKa = 11.84SLSGSVNQTLFRR731 pKa = 5.91
MM1 pKa = 7.75ASILGKK7 pKa = 10.64LGIQEE12 pKa = 4.22GDD14 pKa = 3.58SYY16 pKa = 11.16TAAAGSNRR24 pKa = 11.84AVVVAGTGEE33 pKa = 4.22SAFSSRR39 pKa = 11.84EE40 pKa = 3.51IVGITFGGVAMTRR53 pKa = 11.84AAYY56 pKa = 9.13TGDD59 pKa = 3.4TEE61 pKa = 4.34TAKK64 pKa = 11.02GGVYY68 pKa = 10.35YY69 pKa = 9.99ILEE72 pKa = 4.24SEE74 pKa = 4.38IPAGSSVIAVDD85 pKa = 3.06WDD87 pKa = 4.15GQVHH91 pKa = 6.65DD92 pKa = 5.02AFTGAVYY99 pKa = 9.74TLGGIDD105 pKa = 3.87QVTPVDD111 pKa = 3.93NTNSAIDD118 pKa = 3.52VGQFNLDD125 pKa = 3.15ITFTGVDD132 pKa = 3.48GGVLIVGAGSNSPADD147 pKa = 3.83TLNQSSTISATTTTVTVDD165 pKa = 3.32HH166 pKa = 6.41AQITAGHH173 pKa = 6.52EE174 pKa = 4.18GSAISGVLSAAGSEE188 pKa = 4.43TLGLRR193 pKa = 11.84YY194 pKa = 9.81SSEE197 pKa = 4.03TDD199 pKa = 3.25PVGALAVFNSSGAGPSGPSLDD220 pKa = 4.17TVPSTAYY227 pKa = 9.03PGQSRR232 pKa = 11.84TVSGSGFGATQGTGGLTIGGVSQTITSWSDD262 pKa = 2.83TSITFTAVLGANSYY276 pKa = 11.28GSGKK280 pKa = 8.66TIEE283 pKa = 4.29VTTDD287 pKa = 3.74DD288 pKa = 5.42DD289 pKa = 4.17DD290 pKa = 4.28TDD292 pKa = 3.67SGTIEE297 pKa = 4.7LVADD301 pKa = 3.6TAGGFGVVTMSSPNTADD318 pKa = 3.44EE319 pKa = 4.36SSVAYY324 pKa = 9.88GATPTVVTGDD334 pKa = 3.34QFEE337 pKa = 4.57WEE339 pKa = 4.63DD340 pKa = 4.7FNDD343 pKa = 4.18LGNLAIGSDD352 pKa = 3.73AFVSSVDD359 pKa = 3.39TEE361 pKa = 4.34GTFRR365 pKa = 11.84GRR367 pKa = 11.84FWDD370 pKa = 4.15ASDD373 pKa = 3.99GTWGSVNTFTASATDD388 pKa = 3.5EE389 pKa = 4.21TAPTISSASVPTAGNNIAVQMDD411 pKa = 3.14EE412 pKa = 4.14SMQVGAGGTGGWTISLAGVSVSSASVDD439 pKa = 3.48GSDD442 pKa = 3.39DD443 pKa = 3.65TVVNLTPSRR452 pKa = 11.84TLTDD456 pKa = 3.63EE457 pKa = 4.13DD458 pKa = 4.4TLAIGYY464 pKa = 6.09TQPGDD469 pKa = 3.73GFQDD473 pKa = 3.73QADD476 pKa = 4.38TPNDD480 pKa = 3.51LATLSGQAVTNNSAQEE496 pKa = 4.16PPDD499 pKa = 3.12VTGPVTEE506 pKa = 4.59SVDD509 pKa = 3.53VPTGGTYY516 pKa = 10.4AIGDD520 pKa = 3.94DD521 pKa = 5.22LSFTVNWDD529 pKa = 3.23EE530 pKa = 4.12AVTVTGTPALNLDD543 pKa = 3.09IGGTSRR549 pKa = 11.84QADD552 pKa = 3.93YY553 pKa = 11.45ASGSGSSALVFTYY566 pKa = 9.35TVQSGDD572 pKa = 3.48EE573 pKa = 4.41DD574 pKa = 4.8DD575 pKa = 4.93NGIAVSSLTLEE586 pKa = 4.55GGTLQDD592 pKa = 3.58ASSNNATLTLNSVGDD607 pKa = 3.8TSGVLVDD614 pKa = 4.9GVAPVISINALTTTDD629 pKa = 3.46TTPVVTGSAEE639 pKa = 4.1DD640 pKa = 3.89ADD642 pKa = 4.57SLVLDD647 pKa = 4.66LEE649 pKa = 4.87GVDD652 pKa = 3.55ITHH655 pKa = 6.51SSSYY659 pKa = 10.9NITPSAGSWSQQLPEE674 pKa = 4.37LALGEE679 pKa = 4.2YY680 pKa = 10.79LMTLTGVDD688 pKa = 3.17SAGNEE693 pKa = 4.34TVVTAALNVVEE704 pKa = 5.54SVATSPRR711 pKa = 11.84GLFQPLFRR719 pKa = 11.84SLSGSVNQTLFRR731 pKa = 5.91
Molecular weight: 73.48 kDa
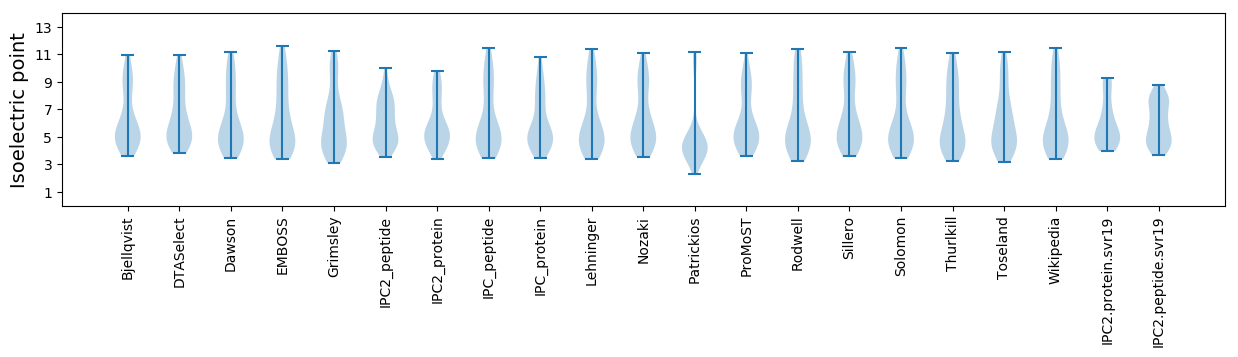
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1GN28|A0A2D1GN28_9CAUD Uncharacterized protein OS=Marinobacter phage PS3 OX=2041341 PE=4 SV=1
MM1 pKa = 7.88DD2 pKa = 5.64RR3 pKa = 11.84FTATEE8 pKa = 4.16FTAINKK14 pKa = 9.79DD15 pKa = 3.16GRR17 pKa = 11.84LTRR20 pKa = 11.84AEE22 pKa = 4.57LEE24 pKa = 4.06TLAWMSEE31 pKa = 3.9GKK33 pKa = 10.12EE34 pKa = 3.76NAAIGILRR42 pKa = 11.84GHH44 pKa = 6.32GTPGAKK50 pKa = 9.94KK51 pKa = 9.98LVSNVMLKK59 pKa = 10.27FNANSRR65 pKa = 11.84CLAIARR71 pKa = 11.84ACRR74 pKa = 11.84DD75 pKa = 3.49GFVVAAAKK83 pKa = 10.04SRR85 pKa = 11.84HH86 pKa = 4.97IAAAFLILASSWVSTTLPDD105 pKa = 3.05SDD107 pKa = 4.07VYY109 pKa = 11.08RR110 pKa = 11.84RR111 pKa = 11.84GANRR115 pKa = 11.84STSVRR120 pKa = 11.84LGPRR124 pKa = 11.84GPGRR128 pKa = 11.84LEE130 pKa = 3.36MDD132 pKa = 4.07LNNWLGAAA140 pKa = 4.27
MM1 pKa = 7.88DD2 pKa = 5.64RR3 pKa = 11.84FTATEE8 pKa = 4.16FTAINKK14 pKa = 9.79DD15 pKa = 3.16GRR17 pKa = 11.84LTRR20 pKa = 11.84AEE22 pKa = 4.57LEE24 pKa = 4.06TLAWMSEE31 pKa = 3.9GKK33 pKa = 10.12EE34 pKa = 3.76NAAIGILRR42 pKa = 11.84GHH44 pKa = 6.32GTPGAKK50 pKa = 9.94KK51 pKa = 9.98LVSNVMLKK59 pKa = 10.27FNANSRR65 pKa = 11.84CLAIARR71 pKa = 11.84ACRR74 pKa = 11.84DD75 pKa = 3.49GFVVAAAKK83 pKa = 10.04SRR85 pKa = 11.84HH86 pKa = 4.97IAAAFLILASSWVSTTLPDD105 pKa = 3.05SDD107 pKa = 4.07VYY109 pKa = 11.08RR110 pKa = 11.84RR111 pKa = 11.84GANRR115 pKa = 11.84STSVRR120 pKa = 11.84LGPRR124 pKa = 11.84GPGRR128 pKa = 11.84LEE130 pKa = 3.36MDD132 pKa = 4.07LNNWLGAAA140 pKa = 4.27
Molecular weight: 15.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12528 |
45 |
911 |
232.0 |
25.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.954 ± 0.482 | 0.894 ± 0.127 |
6.489 ± 0.258 | 6.657 ± 0.272 |
3.52 ± 0.214 | 8.517 ± 0.313 |
1.716 ± 0.185 | 4.829 ± 0.306 |
4.27 ± 0.357 | 8.565 ± 0.264 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.083 ± 0.15 | 3.736 ± 0.19 |
4.502 ± 0.307 | 4.215 ± 0.172 |
5.819 ± 0.414 | 6.992 ± 0.405 |
6.186 ± 0.435 | 6.745 ± 0.301 |
1.557 ± 0.165 | 2.754 ± 0.167 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
