
Aliifodinibius roseus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Aliifodinibius
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
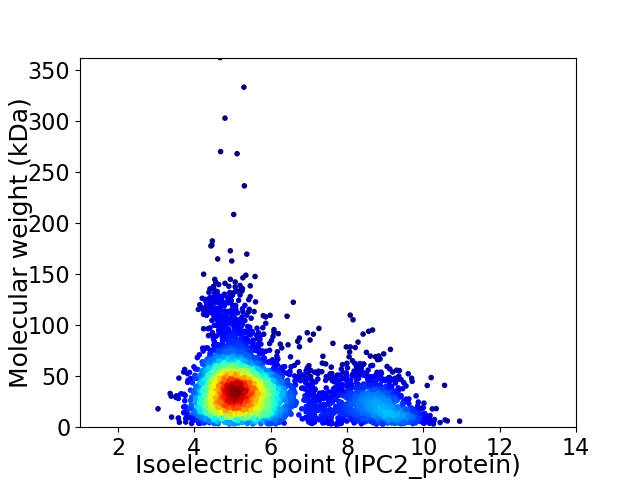
Virtual 2D-PAGE plot for 4289 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5BL67|A0A1M5BL67_9BACT Starch-binding associating with outer membrane OS=Aliifodinibius roseus OX=1194090 GN=SAMN05443144_108164 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 10.56SIVTKK7 pKa = 10.64LIAILMLPSMLLLASCDD24 pKa = 3.44DD25 pKa = 4.59DD26 pKa = 5.81NPVNPEE32 pKa = 3.99PNPPEE37 pKa = 4.64EE38 pKa = 4.11IPAVYY43 pKa = 10.1EE44 pKa = 4.07DD45 pKa = 5.07LPGEE49 pKa = 5.03LISQDD54 pKa = 3.76EE55 pKa = 4.53TWSNDD60 pKa = 2.94TTLTGPRR67 pKa = 11.84FVLPGVTLTVEE78 pKa = 4.16EE79 pKa = 4.43GVEE82 pKa = 4.5VSFTYY87 pKa = 10.5HH88 pKa = 5.55NQNNDD93 pKa = 3.04EE94 pKa = 4.27VGTIITLPGDD104 pKa = 3.32ATNFDD109 pKa = 4.13EE110 pKa = 4.79PRR112 pKa = 11.84ASGRR116 pKa = 11.84LVAVGTADD124 pKa = 3.31NPIIFTAEE132 pKa = 3.95QKK134 pKa = 10.41EE135 pKa = 4.26VASWGGIILAGEE147 pKa = 4.34ASNNVPGGLGEE158 pKa = 5.28IEE160 pKa = 4.67GLDD163 pKa = 3.36QAVQYY168 pKa = 10.65GADD171 pKa = 3.23IGGGEE176 pKa = 4.49SFNDD180 pKa = 3.28QDD182 pKa = 3.89DD183 pKa = 4.14SGRR186 pKa = 11.84LSYY189 pKa = 11.15VRR191 pKa = 11.84IEE193 pKa = 3.87YY194 pKa = 10.15SGYY197 pKa = 10.4SIADD201 pKa = 3.48GSEE204 pKa = 3.86LQALTLYY211 pKa = 10.73SVGSEE216 pKa = 4.06TQLDD220 pKa = 4.28HH221 pKa = 7.79ISIYY225 pKa = 10.59QSVDD229 pKa = 3.09DD230 pKa = 4.28GVEE233 pKa = 4.09LFGGTVDD240 pKa = 4.35IKK242 pKa = 11.2YY243 pKa = 10.55LVIKK247 pKa = 9.68GAQDD251 pKa = 3.15DD252 pKa = 4.36TFDD255 pKa = 4.56YY256 pKa = 11.09DD257 pKa = 3.75QGWTGRR263 pKa = 11.84GQFWVGVQTEE273 pKa = 4.58GTRR276 pKa = 11.84SNSGFEE282 pKa = 4.58NDD284 pKa = 4.37GCDD287 pKa = 4.77DD288 pKa = 4.57QADD291 pKa = 3.94CDD293 pKa = 4.13GGNGPTAPQIYY304 pKa = 9.75NATIYY309 pKa = 11.17GNDD312 pKa = 3.19VANGEE317 pKa = 4.59DD318 pKa = 3.42IRR320 pKa = 11.84GLRR323 pKa = 11.84LRR325 pKa = 11.84EE326 pKa = 4.51GIEE329 pKa = 3.98GEE331 pKa = 4.2YY332 pKa = 10.88KK333 pKa = 10.78NIIIANFGKK342 pKa = 10.42EE343 pKa = 3.79IIAPIVVDD351 pKa = 4.01EE352 pKa = 5.64DD353 pKa = 3.56GTEE356 pKa = 3.97ANIGSSLTFGGNISYY371 pKa = 11.24NNANDD376 pKa = 4.1GSSLSYY382 pKa = 11.13YY383 pKa = 10.73SDD385 pKa = 4.98LGLQNMDD392 pKa = 3.72PQFVDD397 pKa = 2.97AAGFNFALEE406 pKa = 4.29SDD408 pKa = 4.26SPALTSGVAPPVDD421 pKa = 3.98DD422 pKa = 5.87FFDD425 pKa = 3.5QVEE428 pKa = 4.07YY429 pKa = 11.03SGAFGTEE436 pKa = 3.74DD437 pKa = 2.94WTQEE441 pKa = 4.07GSWVRR446 pKa = 11.84WSDD449 pKa = 3.03
MM1 pKa = 7.35KK2 pKa = 10.56SIVTKK7 pKa = 10.64LIAILMLPSMLLLASCDD24 pKa = 3.44DD25 pKa = 4.59DD26 pKa = 5.81NPVNPEE32 pKa = 3.99PNPPEE37 pKa = 4.64EE38 pKa = 4.11IPAVYY43 pKa = 10.1EE44 pKa = 4.07DD45 pKa = 5.07LPGEE49 pKa = 5.03LISQDD54 pKa = 3.76EE55 pKa = 4.53TWSNDD60 pKa = 2.94TTLTGPRR67 pKa = 11.84FVLPGVTLTVEE78 pKa = 4.16EE79 pKa = 4.43GVEE82 pKa = 4.5VSFTYY87 pKa = 10.5HH88 pKa = 5.55NQNNDD93 pKa = 3.04EE94 pKa = 4.27VGTIITLPGDD104 pKa = 3.32ATNFDD109 pKa = 4.13EE110 pKa = 4.79PRR112 pKa = 11.84ASGRR116 pKa = 11.84LVAVGTADD124 pKa = 3.31NPIIFTAEE132 pKa = 3.95QKK134 pKa = 10.41EE135 pKa = 4.26VASWGGIILAGEE147 pKa = 4.34ASNNVPGGLGEE158 pKa = 5.28IEE160 pKa = 4.67GLDD163 pKa = 3.36QAVQYY168 pKa = 10.65GADD171 pKa = 3.23IGGGEE176 pKa = 4.49SFNDD180 pKa = 3.28QDD182 pKa = 3.89DD183 pKa = 4.14SGRR186 pKa = 11.84LSYY189 pKa = 11.15VRR191 pKa = 11.84IEE193 pKa = 3.87YY194 pKa = 10.15SGYY197 pKa = 10.4SIADD201 pKa = 3.48GSEE204 pKa = 3.86LQALTLYY211 pKa = 10.73SVGSEE216 pKa = 4.06TQLDD220 pKa = 4.28HH221 pKa = 7.79ISIYY225 pKa = 10.59QSVDD229 pKa = 3.09DD230 pKa = 4.28GVEE233 pKa = 4.09LFGGTVDD240 pKa = 4.35IKK242 pKa = 11.2YY243 pKa = 10.55LVIKK247 pKa = 9.68GAQDD251 pKa = 3.15DD252 pKa = 4.36TFDD255 pKa = 4.56YY256 pKa = 11.09DD257 pKa = 3.75QGWTGRR263 pKa = 11.84GQFWVGVQTEE273 pKa = 4.58GTRR276 pKa = 11.84SNSGFEE282 pKa = 4.58NDD284 pKa = 4.37GCDD287 pKa = 4.77DD288 pKa = 4.57QADD291 pKa = 3.94CDD293 pKa = 4.13GGNGPTAPQIYY304 pKa = 9.75NATIYY309 pKa = 11.17GNDD312 pKa = 3.19VANGEE317 pKa = 4.59DD318 pKa = 3.42IRR320 pKa = 11.84GLRR323 pKa = 11.84LRR325 pKa = 11.84EE326 pKa = 4.51GIEE329 pKa = 3.98GEE331 pKa = 4.2YY332 pKa = 10.88KK333 pKa = 10.78NIIIANFGKK342 pKa = 10.42EE343 pKa = 3.79IIAPIVVDD351 pKa = 4.01EE352 pKa = 5.64DD353 pKa = 3.56GTEE356 pKa = 3.97ANIGSSLTFGGNISYY371 pKa = 11.24NNANDD376 pKa = 4.1GSSLSYY382 pKa = 11.13YY383 pKa = 10.73SDD385 pKa = 4.98LGLQNMDD392 pKa = 3.72PQFVDD397 pKa = 2.97AAGFNFALEE406 pKa = 4.29SDD408 pKa = 4.26SPALTSGVAPPVDD421 pKa = 3.98DD422 pKa = 5.87FFDD425 pKa = 3.5QVEE428 pKa = 4.07YY429 pKa = 11.03SGAFGTEE436 pKa = 3.74DD437 pKa = 2.94WTQEE441 pKa = 4.07GSWVRR446 pKa = 11.84WSDD449 pKa = 3.03
Molecular weight: 48.11 kDa
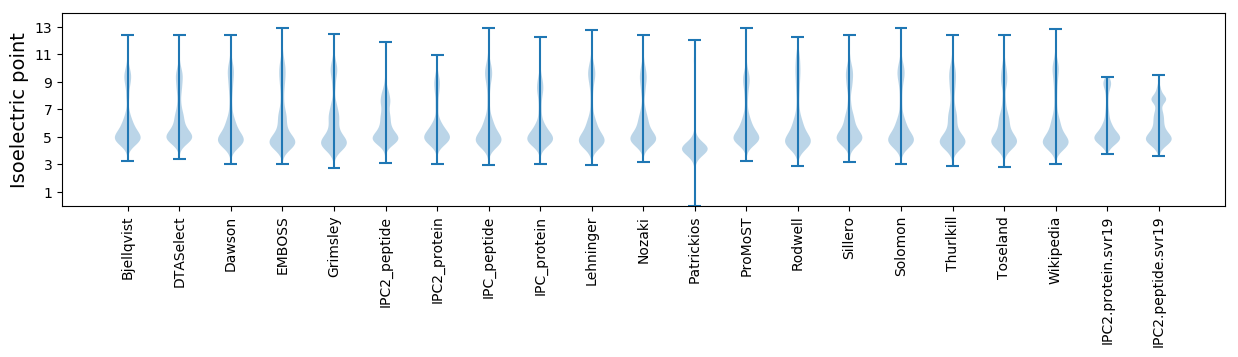
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5ACZ7|A0A1M5ACZ7_9BACT RNA polymerase sigma subunit ECF family OS=Aliifodinibius roseus OX=1194090 GN=SAMN05443144_10728 PE=4 SV=1
MM1 pKa = 7.71WFFLSGFLTPDD12 pKa = 3.51DD13 pKa = 4.36KK14 pKa = 11.72LSFEE18 pKa = 3.9IKK20 pKa = 9.95LRR22 pKa = 11.84QRR24 pKa = 11.84TALVSLLAPSVARR37 pKa = 11.84VFAVLGRR44 pKa = 11.84TYY46 pKa = 11.13SCLSPLLSASRR57 pKa = 11.84CRR59 pKa = 11.84EE60 pKa = 3.62KK61 pKa = 11.22GKK63 pKa = 10.51GRR65 pKa = 11.84NNIPRR70 pKa = 11.84NRR72 pKa = 11.84ICILLLVRR80 pKa = 11.84DD81 pKa = 4.21DD82 pKa = 3.88TAPP85 pKa = 3.24
MM1 pKa = 7.71WFFLSGFLTPDD12 pKa = 3.51DD13 pKa = 4.36KK14 pKa = 11.72LSFEE18 pKa = 3.9IKK20 pKa = 9.95LRR22 pKa = 11.84QRR24 pKa = 11.84TALVSLLAPSVARR37 pKa = 11.84VFAVLGRR44 pKa = 11.84TYY46 pKa = 11.13SCLSPLLSASRR57 pKa = 11.84CRR59 pKa = 11.84EE60 pKa = 3.62KK61 pKa = 11.22GKK63 pKa = 10.51GRR65 pKa = 11.84NNIPRR70 pKa = 11.84NRR72 pKa = 11.84ICILLLVRR80 pKa = 11.84DD81 pKa = 4.21DD82 pKa = 3.88TAPP85 pKa = 3.24
Molecular weight: 9.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1460705 |
28 |
3185 |
340.6 |
38.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.108 ± 0.031 | 0.672 ± 0.011 |
6.057 ± 0.028 | 7.6 ± 0.041 |
4.342 ± 0.024 | 7.459 ± 0.032 |
2.133 ± 0.019 | 6.507 ± 0.034 |
4.993 ± 0.036 | 9.268 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.017 | 4.467 ± 0.032 |
4.24 ± 0.022 | 3.904 ± 0.025 |
5.497 ± 0.028 | 6.489 ± 0.031 |
5.448 ± 0.023 | 6.311 ± 0.03 |
1.356 ± 0.016 | 3.736 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
