
Sporanaerobacter sp. PP17-6a
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellales; Tissierellaceae; Sporanaerobacter; unclassified Sporanaerobacter
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
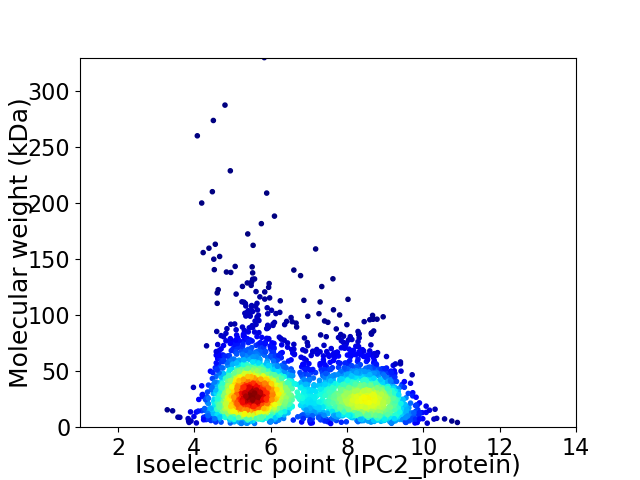
Virtual 2D-PAGE plot for 3134 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4FNG6|A0A1G4FNG6_9FIRM Ribonuclease H OS=Sporanaerobacter sp. PP17-6a OX=1891289 GN=rnhA PE=3 SV=1
MM1 pKa = 7.5GNIALQIEE9 pKa = 4.6RR10 pKa = 11.84TASGATVASTARR22 pKa = 11.84VVFNNLVYY30 pKa = 10.02STGDD34 pKa = 2.72IDD36 pKa = 4.49YY37 pKa = 10.47NPSTGIITLNSTGRR51 pKa = 11.84YY52 pKa = 8.2IINWWVAIQSTGSANGMIFSLLSSQSDD79 pKa = 4.31SITGSSPQIAGQVSGIGIIDD99 pKa = 3.79VVSAPVTLSLINSSTGTLTYY119 pKa = 10.62SVAMPVKK126 pKa = 10.2ASLMVFKK133 pKa = 10.64DD134 pKa = 3.94EE135 pKa = 5.11PSGNSNDD142 pKa = 3.68TTSLCFSYY150 pKa = 10.82AQLAHH155 pKa = 6.74VIEE158 pKa = 4.22QLITIYY164 pKa = 10.37SEE166 pKa = 4.16SVMSVFTTNVNTVTGTPYY184 pKa = 10.77QLYY187 pKa = 10.48ASPDD191 pKa = 3.53GDD193 pKa = 3.27NGAIFVLIDD202 pKa = 3.97SLGQFQAVPITLITAIYY219 pKa = 10.09VGDD222 pKa = 3.54NTVYY226 pKa = 10.85DD227 pKa = 4.15DD228 pKa = 4.81SITYY232 pKa = 7.5LTPPSPLPNGCDD244 pKa = 3.25TDD246 pKa = 4.97LIAAIRR252 pKa = 11.84DD253 pKa = 3.68YY254 pKa = 11.43LPISTEE260 pKa = 4.09VIMQMGATTQASGAIYY276 pKa = 10.06KK277 pKa = 10.08NEE279 pKa = 3.72YY280 pKa = 9.96GILVLSDD287 pKa = 3.66EE288 pKa = 4.86NGNTPIFIPVTKK300 pKa = 9.85IARR303 pKa = 11.84IVTAPPTPSKK313 pKa = 10.82KK314 pKa = 8.27PTPVSVINNPEE325 pKa = 4.17LNVPSNPDD333 pKa = 3.29NPNDD337 pKa = 3.53
MM1 pKa = 7.5GNIALQIEE9 pKa = 4.6RR10 pKa = 11.84TASGATVASTARR22 pKa = 11.84VVFNNLVYY30 pKa = 10.02STGDD34 pKa = 2.72IDD36 pKa = 4.49YY37 pKa = 10.47NPSTGIITLNSTGRR51 pKa = 11.84YY52 pKa = 8.2IINWWVAIQSTGSANGMIFSLLSSQSDD79 pKa = 4.31SITGSSPQIAGQVSGIGIIDD99 pKa = 3.79VVSAPVTLSLINSSTGTLTYY119 pKa = 10.62SVAMPVKK126 pKa = 10.2ASLMVFKK133 pKa = 10.64DD134 pKa = 3.94EE135 pKa = 5.11PSGNSNDD142 pKa = 3.68TTSLCFSYY150 pKa = 10.82AQLAHH155 pKa = 6.74VIEE158 pKa = 4.22QLITIYY164 pKa = 10.37SEE166 pKa = 4.16SVMSVFTTNVNTVTGTPYY184 pKa = 10.77QLYY187 pKa = 10.48ASPDD191 pKa = 3.53GDD193 pKa = 3.27NGAIFVLIDD202 pKa = 3.97SLGQFQAVPITLITAIYY219 pKa = 10.09VGDD222 pKa = 3.54NTVYY226 pKa = 10.85DD227 pKa = 4.15DD228 pKa = 4.81SITYY232 pKa = 7.5LTPPSPLPNGCDD244 pKa = 3.25TDD246 pKa = 4.97LIAAIRR252 pKa = 11.84DD253 pKa = 3.68YY254 pKa = 11.43LPISTEE260 pKa = 4.09VIMQMGATTQASGAIYY276 pKa = 10.06KK277 pKa = 10.08NEE279 pKa = 3.72YY280 pKa = 9.96GILVLSDD287 pKa = 3.66EE288 pKa = 4.86NGNTPIFIPVTKK300 pKa = 9.85IARR303 pKa = 11.84IVTAPPTPSKK313 pKa = 10.82KK314 pKa = 8.27PTPVSVINNPEE325 pKa = 4.17LNVPSNPDD333 pKa = 3.29NPNDD337 pKa = 3.53
Molecular weight: 35.58 kDa
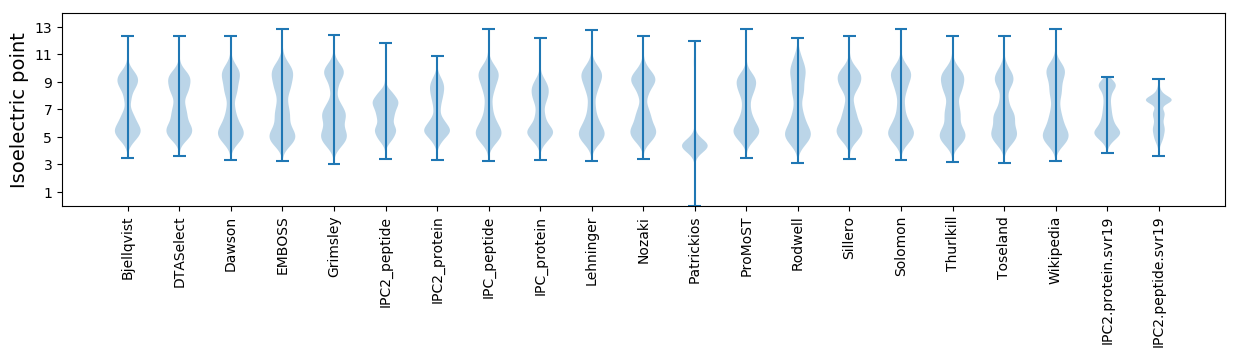
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4FGA3|A0A1G4FGA3_9FIRM Putative D D-dipeptide transport system permease protein DdpC OS=Sporanaerobacter sp. PP17-6a OX=1891289 GN=ddpC PE=3 SV=1
MM1 pKa = 7.64KK2 pKa = 10.42KK3 pKa = 10.18NNKK6 pKa = 9.6LILILTASILSVSMAGCGTKK26 pKa = 10.08TSQRR30 pKa = 11.84PNSRR34 pKa = 11.84VEE36 pKa = 3.89TRR38 pKa = 11.84IGNQAQRR45 pKa = 11.84RR46 pKa = 11.84TPNVMPRR53 pKa = 11.84SNVPRR58 pKa = 11.84LEE60 pKa = 4.32SNLNNNSGIDD70 pKa = 3.65NNGNTNLNTNNNGNMTQGSYY90 pKa = 9.0QRR92 pKa = 11.84AQNIARR98 pKa = 11.84RR99 pKa = 11.84VAALNDD105 pKa = 3.69VNSATAVITGNTALVGVDD123 pKa = 4.82LKK125 pKa = 11.19GTTNDD130 pKa = 3.48NLTTNIRR137 pKa = 11.84NKK139 pKa = 10.08VEE141 pKa = 4.23TAVKK145 pKa = 9.98EE146 pKa = 3.7ADD148 pKa = 3.6KK149 pKa = 11.37NITTVNVTTNPDD161 pKa = 3.27LYY163 pKa = 11.45SRR165 pKa = 11.84ISTIARR171 pKa = 11.84NISNGRR177 pKa = 11.84PMNDD181 pKa = 3.35FANDD185 pKa = 3.09IQDD188 pKa = 3.67ILRR191 pKa = 11.84KK192 pKa = 9.3MNLNRR197 pKa = 4.11
MM1 pKa = 7.64KK2 pKa = 10.42KK3 pKa = 10.18NNKK6 pKa = 9.6LILILTASILSVSMAGCGTKK26 pKa = 10.08TSQRR30 pKa = 11.84PNSRR34 pKa = 11.84VEE36 pKa = 3.89TRR38 pKa = 11.84IGNQAQRR45 pKa = 11.84RR46 pKa = 11.84TPNVMPRR53 pKa = 11.84SNVPRR58 pKa = 11.84LEE60 pKa = 4.32SNLNNNSGIDD70 pKa = 3.65NNGNTNLNTNNNGNMTQGSYY90 pKa = 9.0QRR92 pKa = 11.84AQNIARR98 pKa = 11.84RR99 pKa = 11.84VAALNDD105 pKa = 3.69VNSATAVITGNTALVGVDD123 pKa = 4.82LKK125 pKa = 11.19GTTNDD130 pKa = 3.48NLTTNIRR137 pKa = 11.84NKK139 pKa = 10.08VEE141 pKa = 4.23TAVKK145 pKa = 9.98EE146 pKa = 3.7ADD148 pKa = 3.6KK149 pKa = 11.37NITTVNVTTNPDD161 pKa = 3.27LYY163 pKa = 11.45SRR165 pKa = 11.84ISTIARR171 pKa = 11.84NISNGRR177 pKa = 11.84PMNDD181 pKa = 3.35FANDD185 pKa = 3.09IQDD188 pKa = 3.67ILRR191 pKa = 11.84KK192 pKa = 9.3MNLNRR197 pKa = 4.11
Molecular weight: 21.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
970558 |
29 |
2835 |
309.7 |
35.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.314 ± 0.044 | 1.044 ± 0.017 |
5.693 ± 0.039 | 7.589 ± 0.054 |
4.376 ± 0.032 | 6.788 ± 0.042 |
1.304 ± 0.017 | 10.103 ± 0.048 |
9.044 ± 0.048 | 8.965 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.695 ± 0.019 | 5.904 ± 0.038 |
3.035 ± 0.025 | 2.32 ± 0.02 |
3.755 ± 0.029 | 6.154 ± 0.031 |
4.715 ± 0.029 | 6.357 ± 0.037 |
0.704 ± 0.013 | 4.14 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
