
Pectobacterium phage PPWS1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Corkvirinae; Kotilavirus; Pectobacterium virus PPWS1
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
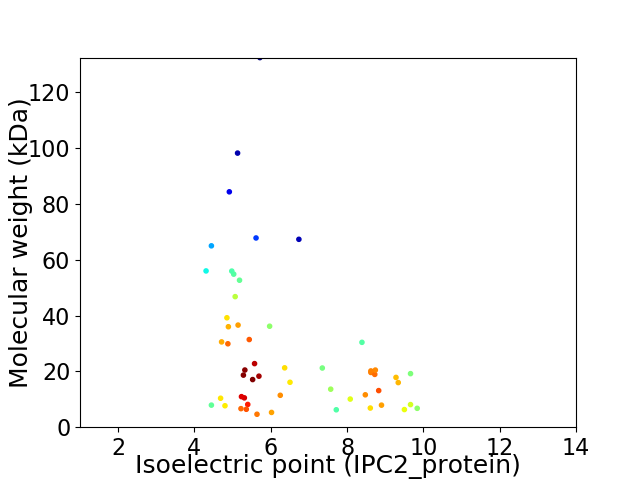
Virtual 2D-PAGE plot for 55 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0UWN1|A0A0P0UWN1_9CAUD Endolysin OS=Pectobacterium phage PPWS1 OX=1685500 GN=56 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.29ISIAEE7 pKa = 4.15PNLSNHH13 pKa = 6.53LEE15 pKa = 4.2GFTARR20 pKa = 11.84VATEE24 pKa = 3.86TRR26 pKa = 11.84SKK28 pKa = 10.76LRR30 pKa = 11.84EE31 pKa = 4.12LFCDD35 pKa = 3.54KK36 pKa = 10.18PASFGVPKK44 pKa = 10.53ALSKK48 pKa = 11.54DD49 pKa = 3.28EE50 pKa = 4.08LTSIKK55 pKa = 10.48QEE57 pKa = 4.0VYY59 pKa = 10.77SRR61 pKa = 11.84ADD63 pKa = 3.1THH65 pKa = 6.16IARR68 pKa = 11.84AVGLLLPPDD77 pKa = 3.58VTIPSAEE84 pKa = 4.08LTDD87 pKa = 3.88DD88 pKa = 5.54ARR90 pKa = 11.84IEE92 pKa = 3.95WMLRR96 pKa = 11.84ALIAAGFAAYY106 pKa = 8.93MRR108 pKa = 11.84EE109 pKa = 4.03AVQDD113 pKa = 3.78YY114 pKa = 10.61SPVLHH119 pKa = 6.08SHH121 pKa = 4.99YY122 pKa = 10.81LEE124 pKa = 4.74YY125 pKa = 11.15ANSWLKK131 pKa = 11.03VFLFKK136 pKa = 11.38NNLPLPRR143 pKa = 11.84SSDD146 pKa = 3.7VASHH150 pKa = 5.52VAVGVAAAPSGTCGYY165 pKa = 10.03IDD167 pKa = 4.18KK168 pKa = 10.8LYY170 pKa = 11.01LRR172 pKa = 11.84LARR175 pKa = 11.84PLVEE179 pKa = 5.74GYY181 pKa = 9.59PPYY184 pKa = 10.42QGLFFQGPTLPPALYY199 pKa = 10.76SLVEE203 pKa = 4.29TNTTAVLATLRR214 pKa = 11.84AEE216 pKa = 4.51MGDD219 pKa = 3.68SPRR222 pKa = 11.84WAVEE226 pKa = 3.43TRR228 pKa = 11.84NFLRR232 pKa = 11.84GVVYY236 pKa = 10.04TEE238 pKa = 4.76AEE240 pKa = 4.01VAMFAGYY247 pKa = 7.8VTLEE251 pKa = 4.13DD252 pKa = 4.63FYY254 pKa = 11.53KK255 pKa = 10.77LNYY258 pKa = 9.39RR259 pKa = 11.84RR260 pKa = 11.84IPLARR265 pKa = 11.84AIKK268 pKa = 9.88EE269 pKa = 3.81ITGSDD274 pKa = 3.13TAAQKK279 pKa = 10.73AADD282 pKa = 3.8VRR284 pKa = 11.84DD285 pKa = 3.69VTSYY289 pKa = 11.43DD290 pKa = 3.29IVLHH294 pKa = 6.14PNDD297 pKa = 3.91RR298 pKa = 11.84PFGAEE303 pKa = 3.7YY304 pKa = 10.97VRR306 pKa = 11.84MRR308 pKa = 11.84EE309 pKa = 4.24EE310 pKa = 3.57NSEE313 pKa = 4.11LHH315 pKa = 6.59SCMTSPAYY323 pKa = 10.04AYY325 pKa = 9.43SSPYY329 pKa = 9.6GVHH332 pKa = 6.44PCDD335 pKa = 3.8VYY337 pKa = 10.19STAYY341 pKa = 9.98YY342 pKa = 10.06GQGDD346 pKa = 3.74NGLVLVEE353 pKa = 4.26ATQFGIPVGRR363 pKa = 11.84GILNVYY369 pKa = 10.39SNTIVRR375 pKa = 11.84WYY377 pKa = 10.05GAYY380 pKa = 9.54KK381 pKa = 10.26AIVMLGNLYY390 pKa = 10.23NIHH393 pKa = 6.29VDD395 pKa = 3.28ADD397 pKa = 3.74SLDD400 pKa = 3.72GVRR403 pKa = 11.84LALIQDD409 pKa = 3.8GDD411 pKa = 5.03LITAPYY417 pKa = 10.54LDD419 pKa = 3.7GCQRR423 pKa = 11.84VEE425 pKa = 4.36LDD427 pKa = 3.84DD428 pKa = 4.32GVLIVDD434 pKa = 3.64ARR436 pKa = 11.84GSIEE440 pKa = 3.89MDD442 pKa = 3.23DD443 pKa = 3.35TSGYY447 pKa = 10.45QEE449 pKa = 3.8TDD451 pKa = 2.86EE452 pKa = 4.19EE453 pKa = 4.39DD454 pKa = 4.02RR455 pKa = 11.84YY456 pKa = 10.99EE457 pKa = 4.08CAISGDD463 pKa = 4.15YY464 pKa = 10.79YY465 pKa = 10.49PEE467 pKa = 4.82SEE469 pKa = 5.19LRR471 pKa = 11.84YY472 pKa = 10.09QSTTDD477 pKa = 2.89TYY479 pKa = 11.57YY480 pKa = 11.3NPDD483 pKa = 3.39NTGMNHH489 pKa = 7.22AEE491 pKa = 4.12HH492 pKa = 6.97CPVLDD497 pKa = 3.61EE498 pKa = 3.85WHH500 pKa = 7.11DD501 pKa = 3.65RR502 pKa = 11.84HH503 pKa = 6.51SGHH506 pKa = 6.92YY507 pKa = 9.82IRR509 pKa = 11.84IDD511 pKa = 3.68GEE513 pKa = 4.39QTWVHH518 pKa = 6.72DD519 pKa = 4.13SVTSDD524 pKa = 3.24EE525 pKa = 5.31LADD528 pKa = 4.65DD529 pKa = 4.59DD530 pKa = 4.75CTYY533 pKa = 10.75MDD535 pKa = 4.17DD536 pKa = 3.94TIGYY540 pKa = 6.44TRR542 pKa = 11.84SPDD545 pKa = 3.43DD546 pKa = 3.97YY547 pKa = 11.65VWDD550 pKa = 4.38DD551 pKa = 3.59VNEE554 pKa = 4.02QYY556 pKa = 9.65YY557 pKa = 8.43TVSDD561 pKa = 3.86YY562 pKa = 11.38DD563 pKa = 3.51YY564 pKa = 10.97LIAEE568 pKa = 4.69RR569 pKa = 11.84EE570 pKa = 4.07QEE572 pKa = 4.0EE573 pKa = 4.79SNEE576 pKa = 4.25SNQQ579 pKa = 3.71
MM1 pKa = 7.64KK2 pKa = 10.29ISIAEE7 pKa = 4.15PNLSNHH13 pKa = 6.53LEE15 pKa = 4.2GFTARR20 pKa = 11.84VATEE24 pKa = 3.86TRR26 pKa = 11.84SKK28 pKa = 10.76LRR30 pKa = 11.84EE31 pKa = 4.12LFCDD35 pKa = 3.54KK36 pKa = 10.18PASFGVPKK44 pKa = 10.53ALSKK48 pKa = 11.54DD49 pKa = 3.28EE50 pKa = 4.08LTSIKK55 pKa = 10.48QEE57 pKa = 4.0VYY59 pKa = 10.77SRR61 pKa = 11.84ADD63 pKa = 3.1THH65 pKa = 6.16IARR68 pKa = 11.84AVGLLLPPDD77 pKa = 3.58VTIPSAEE84 pKa = 4.08LTDD87 pKa = 3.88DD88 pKa = 5.54ARR90 pKa = 11.84IEE92 pKa = 3.95WMLRR96 pKa = 11.84ALIAAGFAAYY106 pKa = 8.93MRR108 pKa = 11.84EE109 pKa = 4.03AVQDD113 pKa = 3.78YY114 pKa = 10.61SPVLHH119 pKa = 6.08SHH121 pKa = 4.99YY122 pKa = 10.81LEE124 pKa = 4.74YY125 pKa = 11.15ANSWLKK131 pKa = 11.03VFLFKK136 pKa = 11.38NNLPLPRR143 pKa = 11.84SSDD146 pKa = 3.7VASHH150 pKa = 5.52VAVGVAAAPSGTCGYY165 pKa = 10.03IDD167 pKa = 4.18KK168 pKa = 10.8LYY170 pKa = 11.01LRR172 pKa = 11.84LARR175 pKa = 11.84PLVEE179 pKa = 5.74GYY181 pKa = 9.59PPYY184 pKa = 10.42QGLFFQGPTLPPALYY199 pKa = 10.76SLVEE203 pKa = 4.29TNTTAVLATLRR214 pKa = 11.84AEE216 pKa = 4.51MGDD219 pKa = 3.68SPRR222 pKa = 11.84WAVEE226 pKa = 3.43TRR228 pKa = 11.84NFLRR232 pKa = 11.84GVVYY236 pKa = 10.04TEE238 pKa = 4.76AEE240 pKa = 4.01VAMFAGYY247 pKa = 7.8VTLEE251 pKa = 4.13DD252 pKa = 4.63FYY254 pKa = 11.53KK255 pKa = 10.77LNYY258 pKa = 9.39RR259 pKa = 11.84RR260 pKa = 11.84IPLARR265 pKa = 11.84AIKK268 pKa = 9.88EE269 pKa = 3.81ITGSDD274 pKa = 3.13TAAQKK279 pKa = 10.73AADD282 pKa = 3.8VRR284 pKa = 11.84DD285 pKa = 3.69VTSYY289 pKa = 11.43DD290 pKa = 3.29IVLHH294 pKa = 6.14PNDD297 pKa = 3.91RR298 pKa = 11.84PFGAEE303 pKa = 3.7YY304 pKa = 10.97VRR306 pKa = 11.84MRR308 pKa = 11.84EE309 pKa = 4.24EE310 pKa = 3.57NSEE313 pKa = 4.11LHH315 pKa = 6.59SCMTSPAYY323 pKa = 10.04AYY325 pKa = 9.43SSPYY329 pKa = 9.6GVHH332 pKa = 6.44PCDD335 pKa = 3.8VYY337 pKa = 10.19STAYY341 pKa = 9.98YY342 pKa = 10.06GQGDD346 pKa = 3.74NGLVLVEE353 pKa = 4.26ATQFGIPVGRR363 pKa = 11.84GILNVYY369 pKa = 10.39SNTIVRR375 pKa = 11.84WYY377 pKa = 10.05GAYY380 pKa = 9.54KK381 pKa = 10.26AIVMLGNLYY390 pKa = 10.23NIHH393 pKa = 6.29VDD395 pKa = 3.28ADD397 pKa = 3.74SLDD400 pKa = 3.72GVRR403 pKa = 11.84LALIQDD409 pKa = 3.8GDD411 pKa = 5.03LITAPYY417 pKa = 10.54LDD419 pKa = 3.7GCQRR423 pKa = 11.84VEE425 pKa = 4.36LDD427 pKa = 3.84DD428 pKa = 4.32GVLIVDD434 pKa = 3.64ARR436 pKa = 11.84GSIEE440 pKa = 3.89MDD442 pKa = 3.23DD443 pKa = 3.35TSGYY447 pKa = 10.45QEE449 pKa = 3.8TDD451 pKa = 2.86EE452 pKa = 4.19EE453 pKa = 4.39DD454 pKa = 4.02RR455 pKa = 11.84YY456 pKa = 10.99EE457 pKa = 4.08CAISGDD463 pKa = 4.15YY464 pKa = 10.79YY465 pKa = 10.49PEE467 pKa = 4.82SEE469 pKa = 5.19LRR471 pKa = 11.84YY472 pKa = 10.09QSTTDD477 pKa = 2.89TYY479 pKa = 11.57YY480 pKa = 11.3NPDD483 pKa = 3.39NTGMNHH489 pKa = 7.22AEE491 pKa = 4.12HH492 pKa = 6.97CPVLDD497 pKa = 3.61EE498 pKa = 3.85WHH500 pKa = 7.11DD501 pKa = 3.65RR502 pKa = 11.84HH503 pKa = 6.51SGHH506 pKa = 6.92YY507 pKa = 9.82IRR509 pKa = 11.84IDD511 pKa = 3.68GEE513 pKa = 4.39QTWVHH518 pKa = 6.72DD519 pKa = 4.13SVTSDD524 pKa = 3.24EE525 pKa = 5.31LADD528 pKa = 4.65DD529 pKa = 4.59DD530 pKa = 4.75CTYY533 pKa = 10.75MDD535 pKa = 4.17DD536 pKa = 3.94TIGYY540 pKa = 6.44TRR542 pKa = 11.84SPDD545 pKa = 3.43DD546 pKa = 3.97YY547 pKa = 11.65VWDD550 pKa = 4.38DD551 pKa = 3.59VNEE554 pKa = 4.02QYY556 pKa = 9.65YY557 pKa = 8.43TVSDD561 pKa = 3.86YY562 pKa = 11.38DD563 pKa = 3.51YY564 pKa = 10.97LIAEE568 pKa = 4.69RR569 pKa = 11.84EE570 pKa = 4.07QEE572 pKa = 4.0EE573 pKa = 4.79SNEE576 pKa = 4.25SNQQ579 pKa = 3.71
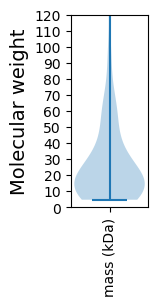
Molecular weight: 65.01 kDa
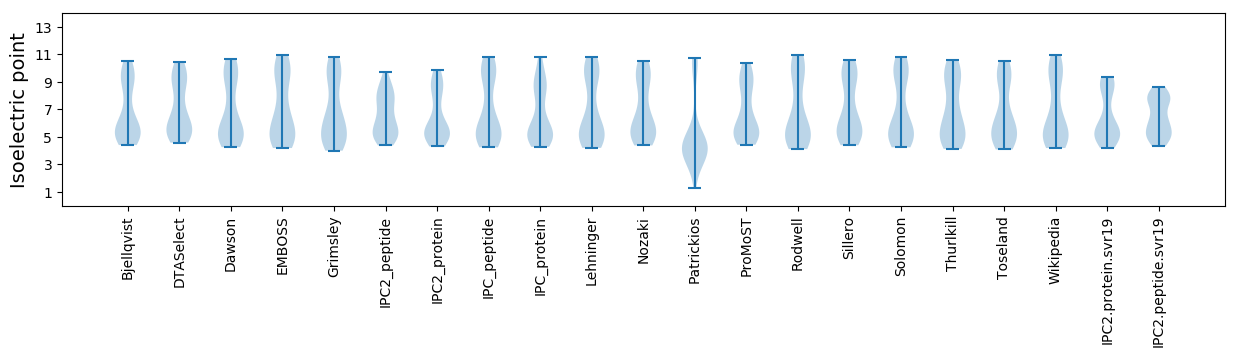
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0UW59|A0A0P0UW59_9CAUD Uncharacterized protein OS=Pectobacterium phage PPWS1 OX=1685500 PE=4 SV=1
MM1 pKa = 7.98EE2 pKa = 4.93IRR4 pKa = 11.84DD5 pKa = 3.99VIAYY9 pKa = 8.92DD10 pKa = 3.52PTSPTGLRR18 pKa = 11.84WLVATSNRR26 pKa = 11.84VKK28 pKa = 10.67VGSPAIRR35 pKa = 11.84AKK37 pKa = 10.75HH38 pKa = 5.22SGGYY42 pKa = 8.27YY43 pKa = 10.08SGMVLGACLEE53 pKa = 4.07PRR55 pKa = 11.84HH56 pKa = 5.68RR57 pKa = 11.84ARR59 pKa = 11.84RR60 pKa = 11.84SGG62 pKa = 3.19
MM1 pKa = 7.98EE2 pKa = 4.93IRR4 pKa = 11.84DD5 pKa = 3.99VIAYY9 pKa = 8.92DD10 pKa = 3.52PTSPTGLRR18 pKa = 11.84WLVATSNRR26 pKa = 11.84VKK28 pKa = 10.67VGSPAIRR35 pKa = 11.84AKK37 pKa = 10.75HH38 pKa = 5.22SGGYY42 pKa = 8.27YY43 pKa = 10.08SGMVLGACLEE53 pKa = 4.07PRR55 pKa = 11.84HH56 pKa = 5.68RR57 pKa = 11.84ARR59 pKa = 11.84RR60 pKa = 11.84SGG62 pKa = 3.19
Molecular weight: 6.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13783 |
43 |
1230 |
250.6 |
27.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.671 ± 0.366 | 1.081 ± 0.173 |
6.392 ± 0.212 | 5.144 ± 0.304 |
2.996 ± 0.159 | 7.596 ± 0.259 |
2.22 ± 0.197 | 4.629 ± 0.166 |
4.041 ± 0.303 | 8.779 ± 0.234 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.822 ± 0.171 | 4.194 ± 0.247 |
4.099 ± 0.228 | 4.484 ± 0.396 |
5.978 ± 0.289 | 6.515 ± 0.294 |
6.624 ± 0.485 | 7.589 ± 0.237 |
1.291 ± 0.134 | 3.853 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
