
Clostridium sp. CAG:568
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
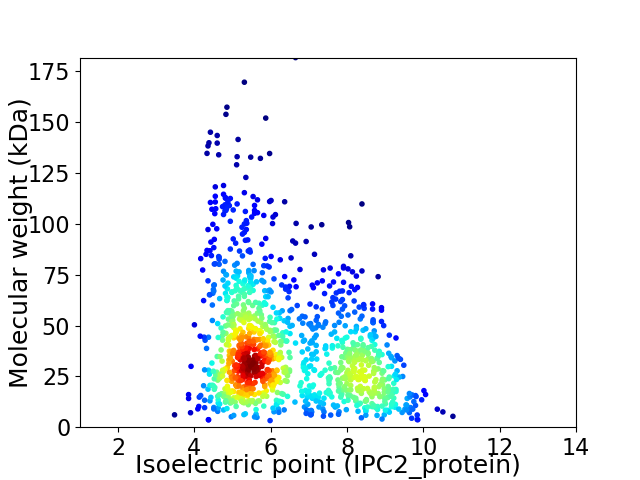
Virtual 2D-PAGE plot for 1211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5ZKK2|R5ZKK2_9CLOT UDP-galactopyranose mutase OS=Clostridium sp. CAG:568 OX=1262821 GN=BN713_00528 PE=3 SV=1
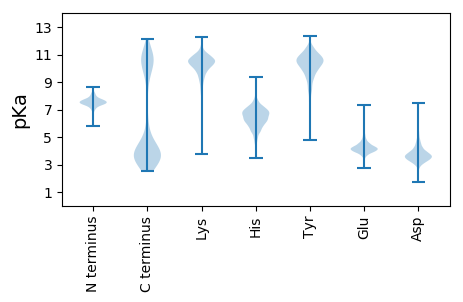
MM1 pKa = 7.13QSKK4 pKa = 10.1FKK6 pKa = 11.07LLTLVLLSTTFFLTSCGDD24 pKa = 3.55STSNPTTPSNQVTDD38 pKa = 3.35NSTNKK43 pKa = 9.82PSAPSVNTDD52 pKa = 3.09KK53 pKa = 11.34KK54 pKa = 10.84DD55 pKa = 3.31TTTSSNKK62 pKa = 9.5KK63 pKa = 10.13DD64 pKa = 3.74DD65 pKa = 4.95ASSDD69 pKa = 2.95WYY71 pKa = 10.84ASEE74 pKa = 4.66KK75 pKa = 10.84EE76 pKa = 3.91IMKK79 pKa = 10.2QAFGFDD85 pKa = 3.52FAIPFADD92 pKa = 3.4GLTDD96 pKa = 3.38NRR98 pKa = 11.84EE99 pKa = 4.0SSLEE103 pKa = 3.94KK104 pKa = 10.46DD105 pKa = 3.16DD106 pKa = 4.79SGNFFYY112 pKa = 10.14THH114 pKa = 7.65DD115 pKa = 4.16MDD117 pKa = 5.6CGNLTSAYY125 pKa = 8.9KK126 pKa = 10.6SKK128 pKa = 11.3VEE130 pKa = 4.02NEE132 pKa = 4.01EE133 pKa = 3.67YY134 pKa = 10.13TYY136 pKa = 11.47DD137 pKa = 3.85STQDD141 pKa = 3.45GYY143 pKa = 11.63DD144 pKa = 3.72LYY146 pKa = 11.1AYY148 pKa = 10.47SYY150 pKa = 11.47DD151 pKa = 3.63DD152 pKa = 3.35TNLIVLQIGYY162 pKa = 9.77SDD164 pKa = 4.6DD165 pKa = 3.88YY166 pKa = 11.73GFEE169 pKa = 3.82IYY171 pKa = 10.33SWVEE175 pKa = 3.88SNTDD179 pKa = 3.54SNVTEE184 pKa = 4.31YY185 pKa = 11.47SSFPYY190 pKa = 10.73ADD192 pKa = 2.85IATFFNLTSVTNNQIPSFDD211 pKa = 3.46IASDD215 pKa = 3.37EE216 pKa = 4.36SYY218 pKa = 11.33YY219 pKa = 11.02GYY221 pKa = 9.39TSKK224 pKa = 10.94SEE226 pKa = 4.04SSSEE230 pKa = 3.69FVVYY234 pKa = 8.14GTIDD238 pKa = 3.24SSITDD243 pKa = 3.31SQMDD247 pKa = 3.9TNYY250 pKa = 11.05SNALKK255 pKa = 10.68ALGYY259 pKa = 8.75TVTVQDD265 pKa = 3.89GYY267 pKa = 11.3YY268 pKa = 9.44PYY270 pKa = 11.13AEE272 pKa = 4.23SEE274 pKa = 4.31EE275 pKa = 4.36KK276 pKa = 10.97GFLLIFGCDD285 pKa = 2.93NSVFFLDD292 pKa = 3.08IMKK295 pKa = 10.4YY296 pKa = 10.1EE297 pKa = 4.33SSTTDD302 pKa = 3.47PDD304 pKa = 4.12TPSTPDD310 pKa = 3.26TTIKK314 pKa = 10.12TITIDD319 pKa = 4.33ANNFDD324 pKa = 3.47AKK326 pKa = 11.1YY327 pKa = 10.01PGFEE331 pKa = 4.17SDD333 pKa = 3.92LTISGYY339 pKa = 7.28TFKK342 pKa = 10.92YY343 pKa = 10.3DD344 pKa = 4.06YY345 pKa = 11.4IMNSNDD351 pKa = 3.25KK352 pKa = 10.52IQFRR356 pKa = 11.84NLSKK360 pKa = 10.99SNSYY364 pKa = 10.56LFNSTAMNLSTVVLNVDD381 pKa = 3.44VTTKK385 pKa = 10.65GDD387 pKa = 3.69YY388 pKa = 10.38VGYY391 pKa = 8.91PSLYY395 pKa = 9.46ASSSILSADD404 pKa = 3.48NNGAEE409 pKa = 4.43IKK411 pKa = 10.45PVITDD416 pKa = 3.35KK417 pKa = 8.88TTYY420 pKa = 10.0TYY422 pKa = 8.94TIPEE426 pKa = 4.3GFSFFKK432 pKa = 10.41VTGHH436 pKa = 6.54KK437 pKa = 10.43NYY439 pKa = 10.88AVLLYY444 pKa = 11.3SMTINLKK451 pKa = 10.28
MM1 pKa = 7.13QSKK4 pKa = 10.1FKK6 pKa = 11.07LLTLVLLSTTFFLTSCGDD24 pKa = 3.55STSNPTTPSNQVTDD38 pKa = 3.35NSTNKK43 pKa = 9.82PSAPSVNTDD52 pKa = 3.09KK53 pKa = 11.34KK54 pKa = 10.84DD55 pKa = 3.31TTTSSNKK62 pKa = 9.5KK63 pKa = 10.13DD64 pKa = 3.74DD65 pKa = 4.95ASSDD69 pKa = 2.95WYY71 pKa = 10.84ASEE74 pKa = 4.66KK75 pKa = 10.84EE76 pKa = 3.91IMKK79 pKa = 10.2QAFGFDD85 pKa = 3.52FAIPFADD92 pKa = 3.4GLTDD96 pKa = 3.38NRR98 pKa = 11.84EE99 pKa = 4.0SSLEE103 pKa = 3.94KK104 pKa = 10.46DD105 pKa = 3.16DD106 pKa = 4.79SGNFFYY112 pKa = 10.14THH114 pKa = 7.65DD115 pKa = 4.16MDD117 pKa = 5.6CGNLTSAYY125 pKa = 8.9KK126 pKa = 10.6SKK128 pKa = 11.3VEE130 pKa = 4.02NEE132 pKa = 4.01EE133 pKa = 3.67YY134 pKa = 10.13TYY136 pKa = 11.47DD137 pKa = 3.85STQDD141 pKa = 3.45GYY143 pKa = 11.63DD144 pKa = 3.72LYY146 pKa = 11.1AYY148 pKa = 10.47SYY150 pKa = 11.47DD151 pKa = 3.63DD152 pKa = 3.35TNLIVLQIGYY162 pKa = 9.77SDD164 pKa = 4.6DD165 pKa = 3.88YY166 pKa = 11.73GFEE169 pKa = 3.82IYY171 pKa = 10.33SWVEE175 pKa = 3.88SNTDD179 pKa = 3.54SNVTEE184 pKa = 4.31YY185 pKa = 11.47SSFPYY190 pKa = 10.73ADD192 pKa = 2.85IATFFNLTSVTNNQIPSFDD211 pKa = 3.46IASDD215 pKa = 3.37EE216 pKa = 4.36SYY218 pKa = 11.33YY219 pKa = 11.02GYY221 pKa = 9.39TSKK224 pKa = 10.94SEE226 pKa = 4.04SSSEE230 pKa = 3.69FVVYY234 pKa = 8.14GTIDD238 pKa = 3.24SSITDD243 pKa = 3.31SQMDD247 pKa = 3.9TNYY250 pKa = 11.05SNALKK255 pKa = 10.68ALGYY259 pKa = 8.75TVTVQDD265 pKa = 3.89GYY267 pKa = 11.3YY268 pKa = 9.44PYY270 pKa = 11.13AEE272 pKa = 4.23SEE274 pKa = 4.31EE275 pKa = 4.36KK276 pKa = 10.97GFLLIFGCDD285 pKa = 2.93NSVFFLDD292 pKa = 3.08IMKK295 pKa = 10.4YY296 pKa = 10.1EE297 pKa = 4.33SSTTDD302 pKa = 3.47PDD304 pKa = 4.12TPSTPDD310 pKa = 3.26TTIKK314 pKa = 10.12TITIDD319 pKa = 4.33ANNFDD324 pKa = 3.47AKK326 pKa = 11.1YY327 pKa = 10.01PGFEE331 pKa = 4.17SDD333 pKa = 3.92LTISGYY339 pKa = 7.28TFKK342 pKa = 10.92YY343 pKa = 10.3DD344 pKa = 4.06YY345 pKa = 11.4IMNSNDD351 pKa = 3.25KK352 pKa = 10.52IQFRR356 pKa = 11.84NLSKK360 pKa = 10.99SNSYY364 pKa = 10.56LFNSTAMNLSTVVLNVDD381 pKa = 3.44VTTKK385 pKa = 10.65GDD387 pKa = 3.69YY388 pKa = 10.38VGYY391 pKa = 8.91PSLYY395 pKa = 9.46ASSSILSADD404 pKa = 3.48NNGAEE409 pKa = 4.43IKK411 pKa = 10.45PVITDD416 pKa = 3.35KK417 pKa = 8.88TTYY420 pKa = 10.0TYY422 pKa = 8.94TIPEE426 pKa = 4.3GFSFFKK432 pKa = 10.41VTGHH436 pKa = 6.54KK437 pKa = 10.43NYY439 pKa = 10.88AVLLYY444 pKa = 11.3SMTINLKK451 pKa = 10.28
Molecular weight: 50.36 kDa
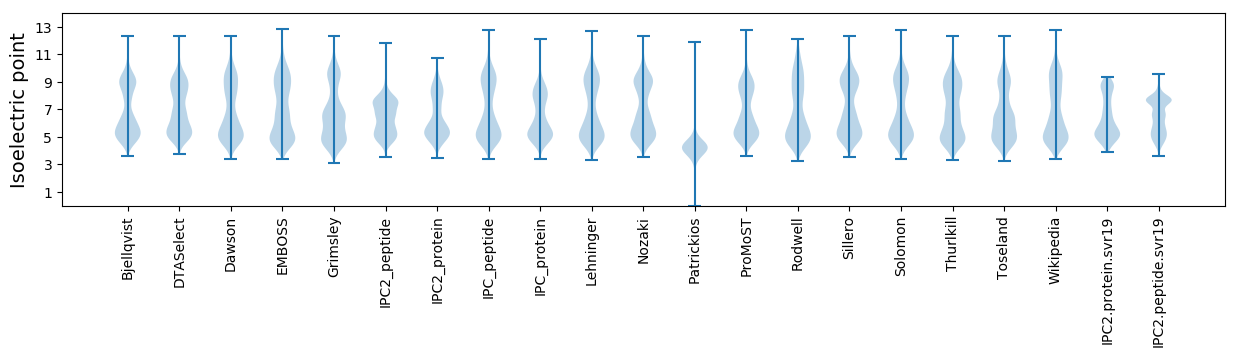
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5ZTC5|R5ZTC5_9CLOT HATPase_c_5 domain-containing protein OS=Clostridium sp. CAG:568 OX=1262821 GN=BN713_01102 PE=4 SV=1
MM1 pKa = 7.53AVPTRR6 pKa = 11.84RR7 pKa = 11.84ASKK10 pKa = 7.97TRR12 pKa = 11.84KK13 pKa = 8.94RR14 pKa = 11.84LRR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 6.21FHH21 pKa = 6.74LAEE24 pKa = 4.1QNLTRR29 pKa = 11.84CSNCGAMIQSHH40 pKa = 7.06RR41 pKa = 11.84ICSSCGFYY49 pKa = 10.23RR50 pKa = 11.84GKK52 pKa = 10.49KK53 pKa = 9.08VLDD56 pKa = 3.58VKK58 pKa = 11.12VKK60 pKa = 10.58EE61 pKa = 4.34DD62 pKa = 3.19
MM1 pKa = 7.53AVPTRR6 pKa = 11.84RR7 pKa = 11.84ASKK10 pKa = 7.97TRR12 pKa = 11.84KK13 pKa = 8.94RR14 pKa = 11.84LRR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 6.21FHH21 pKa = 6.74LAEE24 pKa = 4.1QNLTRR29 pKa = 11.84CSNCGAMIQSHH40 pKa = 7.06RR41 pKa = 11.84ICSSCGFYY49 pKa = 10.23RR50 pKa = 11.84GKK52 pKa = 10.49KK53 pKa = 9.08VLDD56 pKa = 3.58VKK58 pKa = 11.12VKK60 pKa = 10.58EE61 pKa = 4.34DD62 pKa = 3.19
Molecular weight: 7.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
438710 |
30 |
1570 |
362.3 |
40.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.659 ± 0.068 | 1.177 ± 0.022 |
6.751 ± 0.054 | 6.346 ± 0.066 |
4.863 ± 0.056 | 6.292 ± 0.056 |
1.439 ± 0.026 | 7.921 ± 0.075 |
8.649 ± 0.073 | 8.896 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.472 ± 0.032 | 5.939 ± 0.053 |
3.087 ± 0.041 | 2.553 ± 0.033 |
3.348 ± 0.05 | 7.413 ± 0.074 |
5.571 ± 0.081 | 6.229 ± 0.051 |
0.798 ± 0.023 | 4.593 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
