
Acinetobacter phage AP205
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Leviviricetes; Norzivirales; Duinviridae; Apeevirus; Apeevirus quebecense
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
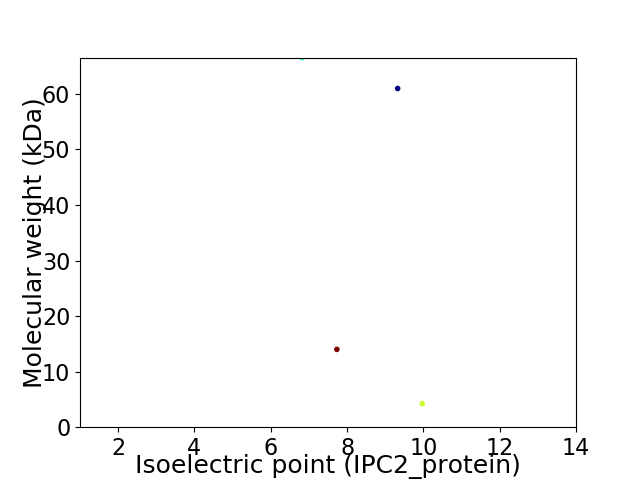
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
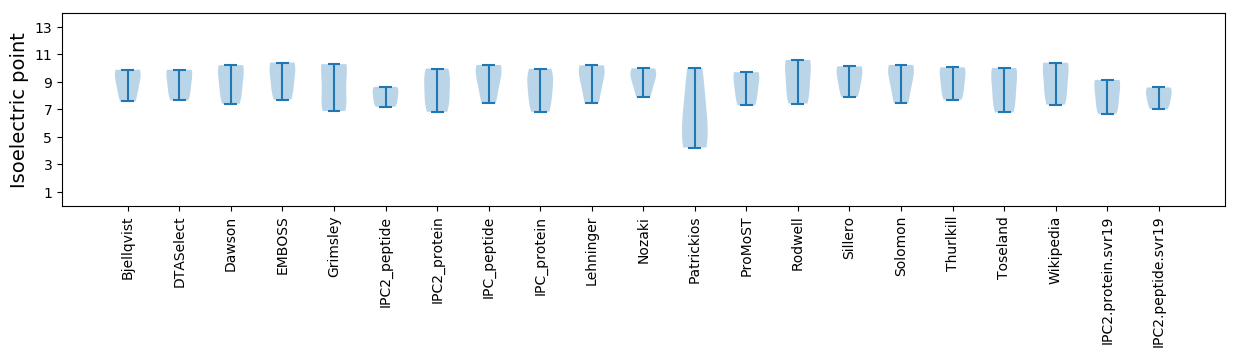
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9AZ42|Q9AZ42_9VIRU Coat protein OS=Acinetobacter phage AP205 OX=154784 PE=1 SV=1
MM1 pKa = 7.72LNACDD6 pKa = 3.9QLKK9 pKa = 8.86STSIPAFQSNVLSDD23 pKa = 3.84VLSLSDD29 pKa = 3.98AADD32 pKa = 3.01ITVKK36 pKa = 10.44HH37 pKa = 6.31RR38 pKa = 11.84IVSKK42 pKa = 10.32FGEE45 pKa = 4.28PAGSSLRR52 pKa = 11.84DD53 pKa = 3.31VAFNNYY59 pKa = 9.75KK60 pKa = 10.54LFEE63 pKa = 4.05QHH65 pKa = 6.57LGSIPQITNLWQEE78 pKa = 3.83GKK80 pKa = 10.64EE81 pKa = 3.97FFFLRR86 pKa = 11.84KK87 pKa = 9.76AKK89 pKa = 10.77ANLGKK94 pKa = 9.44WLKK97 pKa = 9.16TFKK100 pKa = 10.64LDD102 pKa = 3.64YY103 pKa = 11.07NSITVEE109 pKa = 4.0FTPGEE114 pKa = 4.57SYY116 pKa = 10.86TSATGHH122 pKa = 5.39VSVFAKK128 pKa = 10.53LSNLAHH134 pKa = 5.93WTCTADD140 pKa = 4.03VVDD143 pKa = 5.09DD144 pKa = 4.1VCHH147 pKa = 5.67LVYY150 pKa = 10.93YY151 pKa = 10.81NRR153 pKa = 11.84GLKK156 pKa = 9.71AAARR160 pKa = 11.84KK161 pKa = 9.57HH162 pKa = 5.68IGLMVPIEE170 pKa = 4.41GEE172 pKa = 4.11SGFDD176 pKa = 3.02TFSRR180 pKa = 11.84HH181 pKa = 4.55LMGVISIVPGARR193 pKa = 11.84GASVPKK199 pKa = 10.36NQEE202 pKa = 3.04TDD204 pKa = 2.9RR205 pKa = 11.84FIDD208 pKa = 3.58VEE210 pKa = 4.29PTFNMILQRR219 pKa = 11.84WVAGEE224 pKa = 3.62ITRR227 pKa = 11.84CLTLAKK233 pKa = 10.04NHH235 pKa = 6.44LGASRR240 pKa = 11.84NINGKK245 pKa = 8.33VVFHH249 pKa = 7.44DD250 pKa = 3.47AQEE253 pKa = 4.03LHH255 pKa = 7.02KK256 pKa = 11.09EE257 pKa = 4.17MIRR260 pKa = 11.84DD261 pKa = 3.93LSYY264 pKa = 10.82ATIDD268 pKa = 3.6FSNASDD274 pKa = 4.67SVLLWVVQLLFPKK287 pKa = 10.03HH288 pKa = 5.2VSYY291 pKa = 11.57VLTQYY296 pKa = 10.82RR297 pKa = 11.84SSTVQLGSDD306 pKa = 4.1LIEE309 pKa = 4.53PNKK312 pKa = 10.03LSSMGNGFTFEE323 pKa = 4.2VMTLLLLSIGRR334 pKa = 11.84IFDD337 pKa = 3.56PTCRR341 pKa = 11.84VYY343 pKa = 11.34GDD345 pKa = 3.84DD346 pKa = 4.19VIIKK350 pKa = 10.65AEE352 pKa = 4.04VADD355 pKa = 4.4DD356 pKa = 4.65FINTVSSIAFMTNNKK371 pKa = 8.38KK372 pKa = 8.16TFLKK376 pKa = 10.7GLFRR380 pKa = 11.84EE381 pKa = 4.49SCGAFQFDD389 pKa = 3.82TFDD392 pKa = 3.23IQSFEE397 pKa = 4.69FEE399 pKa = 4.04WADD402 pKa = 3.35NFTDD406 pKa = 5.81VIAICNKK413 pKa = 9.96LKK415 pKa = 10.98LIIDD419 pKa = 4.49AAQCNEE425 pKa = 3.6AVIAILRR432 pKa = 11.84NAHH435 pKa = 5.55TVICEE440 pKa = 4.43CIPVLCKK447 pKa = 10.62GPQPPDD453 pKa = 3.41FNLFLSQYY461 pKa = 10.75VYY463 pKa = 11.43DD464 pKa = 5.08DD465 pKa = 3.2NWKK468 pKa = 9.4KK469 pKa = 10.97KK470 pKa = 7.46QMKK473 pKa = 9.85SDD475 pKa = 3.74LAITKK480 pKa = 10.11LNRR483 pKa = 11.84LVDD486 pKa = 3.89KK487 pKa = 10.97QWGFFSATHH496 pKa = 5.57HH497 pKa = 6.52HH498 pKa = 7.09PEE500 pKa = 3.84EE501 pKa = 4.18LCYY504 pKa = 11.18VNIPVYY510 pKa = 10.26VPRR513 pKa = 11.84RR514 pKa = 11.84DD515 pKa = 3.63SVHH518 pKa = 6.7AGQNLFVDD526 pKa = 4.8LSNLYY531 pKa = 10.55ALRR534 pKa = 11.84FTKK537 pKa = 9.8STVRR541 pKa = 11.84GKK543 pKa = 10.73GKK545 pKa = 9.22WVNVPHH551 pKa = 7.02WVTPVGSIYY560 pKa = 9.84RR561 pKa = 11.84ASRR564 pKa = 11.84IRR566 pKa = 11.84QQYY569 pKa = 8.68PNIGEE574 pKa = 4.91LPTCYY579 pKa = 9.69WSPHH583 pKa = 4.8QLDD586 pKa = 5.71LITSS590 pKa = 4.18
MM1 pKa = 7.72LNACDD6 pKa = 3.9QLKK9 pKa = 8.86STSIPAFQSNVLSDD23 pKa = 3.84VLSLSDD29 pKa = 3.98AADD32 pKa = 3.01ITVKK36 pKa = 10.44HH37 pKa = 6.31RR38 pKa = 11.84IVSKK42 pKa = 10.32FGEE45 pKa = 4.28PAGSSLRR52 pKa = 11.84DD53 pKa = 3.31VAFNNYY59 pKa = 9.75KK60 pKa = 10.54LFEE63 pKa = 4.05QHH65 pKa = 6.57LGSIPQITNLWQEE78 pKa = 3.83GKK80 pKa = 10.64EE81 pKa = 3.97FFFLRR86 pKa = 11.84KK87 pKa = 9.76AKK89 pKa = 10.77ANLGKK94 pKa = 9.44WLKK97 pKa = 9.16TFKK100 pKa = 10.64LDD102 pKa = 3.64YY103 pKa = 11.07NSITVEE109 pKa = 4.0FTPGEE114 pKa = 4.57SYY116 pKa = 10.86TSATGHH122 pKa = 5.39VSVFAKK128 pKa = 10.53LSNLAHH134 pKa = 5.93WTCTADD140 pKa = 4.03VVDD143 pKa = 5.09DD144 pKa = 4.1VCHH147 pKa = 5.67LVYY150 pKa = 10.93YY151 pKa = 10.81NRR153 pKa = 11.84GLKK156 pKa = 9.71AAARR160 pKa = 11.84KK161 pKa = 9.57HH162 pKa = 5.68IGLMVPIEE170 pKa = 4.41GEE172 pKa = 4.11SGFDD176 pKa = 3.02TFSRR180 pKa = 11.84HH181 pKa = 4.55LMGVISIVPGARR193 pKa = 11.84GASVPKK199 pKa = 10.36NQEE202 pKa = 3.04TDD204 pKa = 2.9RR205 pKa = 11.84FIDD208 pKa = 3.58VEE210 pKa = 4.29PTFNMILQRR219 pKa = 11.84WVAGEE224 pKa = 3.62ITRR227 pKa = 11.84CLTLAKK233 pKa = 10.04NHH235 pKa = 6.44LGASRR240 pKa = 11.84NINGKK245 pKa = 8.33VVFHH249 pKa = 7.44DD250 pKa = 3.47AQEE253 pKa = 4.03LHH255 pKa = 7.02KK256 pKa = 11.09EE257 pKa = 4.17MIRR260 pKa = 11.84DD261 pKa = 3.93LSYY264 pKa = 10.82ATIDD268 pKa = 3.6FSNASDD274 pKa = 4.67SVLLWVVQLLFPKK287 pKa = 10.03HH288 pKa = 5.2VSYY291 pKa = 11.57VLTQYY296 pKa = 10.82RR297 pKa = 11.84SSTVQLGSDD306 pKa = 4.1LIEE309 pKa = 4.53PNKK312 pKa = 10.03LSSMGNGFTFEE323 pKa = 4.2VMTLLLLSIGRR334 pKa = 11.84IFDD337 pKa = 3.56PTCRR341 pKa = 11.84VYY343 pKa = 11.34GDD345 pKa = 3.84DD346 pKa = 4.19VIIKK350 pKa = 10.65AEE352 pKa = 4.04VADD355 pKa = 4.4DD356 pKa = 4.65FINTVSSIAFMTNNKK371 pKa = 8.38KK372 pKa = 8.16TFLKK376 pKa = 10.7GLFRR380 pKa = 11.84EE381 pKa = 4.49SCGAFQFDD389 pKa = 3.82TFDD392 pKa = 3.23IQSFEE397 pKa = 4.69FEE399 pKa = 4.04WADD402 pKa = 3.35NFTDD406 pKa = 5.81VIAICNKK413 pKa = 9.96LKK415 pKa = 10.98LIIDD419 pKa = 4.49AAQCNEE425 pKa = 3.6AVIAILRR432 pKa = 11.84NAHH435 pKa = 5.55TVICEE440 pKa = 4.43CIPVLCKK447 pKa = 10.62GPQPPDD453 pKa = 3.41FNLFLSQYY461 pKa = 10.75VYY463 pKa = 11.43DD464 pKa = 5.08DD465 pKa = 3.2NWKK468 pKa = 9.4KK469 pKa = 10.97KK470 pKa = 7.46QMKK473 pKa = 9.85SDD475 pKa = 3.74LAITKK480 pKa = 10.11LNRR483 pKa = 11.84LVDD486 pKa = 3.89KK487 pKa = 10.97QWGFFSATHH496 pKa = 5.57HH497 pKa = 6.52HH498 pKa = 7.09PEE500 pKa = 3.84EE501 pKa = 4.18LCYY504 pKa = 11.18VNIPVYY510 pKa = 10.26VPRR513 pKa = 11.84RR514 pKa = 11.84DD515 pKa = 3.63SVHH518 pKa = 6.7AGQNLFVDD526 pKa = 4.8LSNLYY531 pKa = 10.55ALRR534 pKa = 11.84FTKK537 pKa = 9.8STVRR541 pKa = 11.84GKK543 pKa = 10.73GKK545 pKa = 9.22WVNVPHH551 pKa = 7.02WVTPVGSIYY560 pKa = 9.84RR561 pKa = 11.84ASRR564 pKa = 11.84IRR566 pKa = 11.84QQYY569 pKa = 8.68PNIGEE574 pKa = 4.91LPTCYY579 pKa = 9.69WSPHH583 pKa = 4.8QLDD586 pKa = 5.71LITSS590 pKa = 4.18
Molecular weight: 66.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9AZ45|Q9AZ45_9VIRU Lysis protein OS=Acinetobacter phage AP205 OX=154784 PE=4 SV=1
MM1 pKa = 7.39NMYY4 pKa = 9.94KK5 pKa = 9.81WVPEE9 pKa = 4.24SIRR12 pKa = 11.84DD13 pKa = 3.66SGEE16 pKa = 3.87GQPSYY21 pKa = 11.61SNNGDD26 pKa = 3.77YY27 pKa = 10.89APSGPWVAAGIHH39 pKa = 5.7TMPQSLRR46 pKa = 11.84DD47 pKa = 3.63SMRR50 pKa = 11.84NSIMVTAQARR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.76VIGPEE67 pKa = 3.67WGPDD71 pKa = 3.27GRR73 pKa = 11.84FTGYY77 pKa = 10.83ASVIGTPDD85 pKa = 3.92PKK87 pKa = 10.65PADD90 pKa = 3.27IVNKK94 pKa = 8.17FTVEE98 pKa = 3.69RR99 pKa = 11.84RR100 pKa = 11.84PVSNGNFQQRR110 pKa = 11.84VKK112 pKa = 11.07AGDD115 pKa = 3.48IVVAPYY121 pKa = 10.31TSDD124 pKa = 2.99GKK126 pKa = 9.55ITVKK130 pKa = 10.5LVAGQKK136 pKa = 10.14DD137 pKa = 3.55ISSTPDD143 pKa = 2.86YY144 pKa = 10.68DD145 pKa = 3.68YY146 pKa = 11.21RR147 pKa = 11.84IDD149 pKa = 3.74SSLASSAGFVVAGEE163 pKa = 3.62RR164 pKa = 11.84WYY166 pKa = 8.65YY167 pKa = 8.15TKK169 pKa = 10.6RR170 pKa = 11.84HH171 pKa = 6.14FIIPRR176 pKa = 11.84YY177 pKa = 7.01FQNWRR182 pKa = 11.84MRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84KK187 pKa = 9.79YY188 pKa = 8.05VTGWVMPTFYY198 pKa = 10.71SPKK201 pKa = 10.09EE202 pKa = 3.51IFNRR206 pKa = 11.84LKK208 pKa = 10.93DD209 pKa = 3.65SLVPDD214 pKa = 3.99TGLVTQVWADD224 pKa = 3.54NNTKK228 pKa = 10.58RR229 pKa = 11.84MDD231 pKa = 3.69FLTAMAEE238 pKa = 4.18IPQTLSSFLDD248 pKa = 3.23ALGYY252 pKa = 10.31LGSLIKK258 pKa = 10.54DD259 pKa = 4.05FKK261 pKa = 10.69RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84FFLNKK269 pKa = 8.26AHH271 pKa = 6.24QRR273 pKa = 11.84IRR275 pKa = 11.84NKK277 pKa = 10.54LGVSFAEE284 pKa = 3.87RR285 pKa = 11.84RR286 pKa = 11.84SQIVSKK292 pKa = 10.22YY293 pKa = 9.33DD294 pKa = 3.28RR295 pKa = 11.84KK296 pKa = 10.03IASARR301 pKa = 11.84KK302 pKa = 7.89PAIIVKK308 pKa = 9.84LRR310 pKa = 11.84QRR312 pKa = 11.84KK313 pKa = 8.38EE314 pKa = 3.83KK315 pKa = 10.57ALKK318 pKa = 10.51ALDD321 pKa = 3.35KK322 pKa = 10.69MRR324 pKa = 11.84VRR326 pKa = 11.84EE327 pKa = 4.19EE328 pKa = 3.49KK329 pKa = 10.92KK330 pKa = 9.89MIRR333 pKa = 11.84EE334 pKa = 4.27FATQAASLWLSFRR347 pKa = 11.84YY348 pKa = 9.84EE349 pKa = 4.0IMPLYY354 pKa = 10.12YY355 pKa = 10.08QSQDD359 pKa = 3.51VLDD362 pKa = 4.87VIANSTSEE370 pKa = 4.05FMTSRR375 pKa = 11.84DD376 pKa = 3.65FVAKK380 pKa = 10.13AINIGIPLEE389 pKa = 4.14WNLDD393 pKa = 3.57QEE395 pKa = 4.78NLVSQPRR402 pKa = 11.84HH403 pKa = 4.02NVMVKK408 pKa = 10.43SKK410 pKa = 10.74LSPEE414 pKa = 3.91NNIGKK419 pKa = 7.59TLSVNPFTTAWEE431 pKa = 4.11LLTLSFVVDD440 pKa = 3.13WFVNFGDD447 pKa = 4.23VIAGFTGGYY456 pKa = 9.51SDD458 pKa = 6.04DD459 pKa = 4.26SGATASWRR467 pKa = 11.84FDD469 pKa = 3.61DD470 pKa = 5.31KK471 pKa = 11.53KK472 pKa = 11.25VFHH475 pKa = 6.96LKK477 pKa = 10.34NIPSAMVIVDD487 pKa = 2.93INFYY491 pKa = 8.72TRR493 pKa = 11.84QVIDD497 pKa = 3.59PRR499 pKa = 11.84LCGGLAFSPKK509 pKa = 10.36LNLFRR514 pKa = 11.84YY515 pKa = 9.22LDD517 pKa = 3.93AMSLSWNRR525 pKa = 11.84SRR527 pKa = 11.84LKK529 pKa = 10.38ISRR532 pKa = 11.84ATT534 pKa = 3.32
MM1 pKa = 7.39NMYY4 pKa = 9.94KK5 pKa = 9.81WVPEE9 pKa = 4.24SIRR12 pKa = 11.84DD13 pKa = 3.66SGEE16 pKa = 3.87GQPSYY21 pKa = 11.61SNNGDD26 pKa = 3.77YY27 pKa = 10.89APSGPWVAAGIHH39 pKa = 5.7TMPQSLRR46 pKa = 11.84DD47 pKa = 3.63SMRR50 pKa = 11.84NSIMVTAQARR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.76VIGPEE67 pKa = 3.67WGPDD71 pKa = 3.27GRR73 pKa = 11.84FTGYY77 pKa = 10.83ASVIGTPDD85 pKa = 3.92PKK87 pKa = 10.65PADD90 pKa = 3.27IVNKK94 pKa = 8.17FTVEE98 pKa = 3.69RR99 pKa = 11.84RR100 pKa = 11.84PVSNGNFQQRR110 pKa = 11.84VKK112 pKa = 11.07AGDD115 pKa = 3.48IVVAPYY121 pKa = 10.31TSDD124 pKa = 2.99GKK126 pKa = 9.55ITVKK130 pKa = 10.5LVAGQKK136 pKa = 10.14DD137 pKa = 3.55ISSTPDD143 pKa = 2.86YY144 pKa = 10.68DD145 pKa = 3.68YY146 pKa = 11.21RR147 pKa = 11.84IDD149 pKa = 3.74SSLASSAGFVVAGEE163 pKa = 3.62RR164 pKa = 11.84WYY166 pKa = 8.65YY167 pKa = 8.15TKK169 pKa = 10.6RR170 pKa = 11.84HH171 pKa = 6.14FIIPRR176 pKa = 11.84YY177 pKa = 7.01FQNWRR182 pKa = 11.84MRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84KK187 pKa = 9.79YY188 pKa = 8.05VTGWVMPTFYY198 pKa = 10.71SPKK201 pKa = 10.09EE202 pKa = 3.51IFNRR206 pKa = 11.84LKK208 pKa = 10.93DD209 pKa = 3.65SLVPDD214 pKa = 3.99TGLVTQVWADD224 pKa = 3.54NNTKK228 pKa = 10.58RR229 pKa = 11.84MDD231 pKa = 3.69FLTAMAEE238 pKa = 4.18IPQTLSSFLDD248 pKa = 3.23ALGYY252 pKa = 10.31LGSLIKK258 pKa = 10.54DD259 pKa = 4.05FKK261 pKa = 10.69RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84FFLNKK269 pKa = 8.26AHH271 pKa = 6.24QRR273 pKa = 11.84IRR275 pKa = 11.84NKK277 pKa = 10.54LGVSFAEE284 pKa = 3.87RR285 pKa = 11.84RR286 pKa = 11.84SQIVSKK292 pKa = 10.22YY293 pKa = 9.33DD294 pKa = 3.28RR295 pKa = 11.84KK296 pKa = 10.03IASARR301 pKa = 11.84KK302 pKa = 7.89PAIIVKK308 pKa = 9.84LRR310 pKa = 11.84QRR312 pKa = 11.84KK313 pKa = 8.38EE314 pKa = 3.83KK315 pKa = 10.57ALKK318 pKa = 10.51ALDD321 pKa = 3.35KK322 pKa = 10.69MRR324 pKa = 11.84VRR326 pKa = 11.84EE327 pKa = 4.19EE328 pKa = 3.49KK329 pKa = 10.92KK330 pKa = 9.89MIRR333 pKa = 11.84EE334 pKa = 4.27FATQAASLWLSFRR347 pKa = 11.84YY348 pKa = 9.84EE349 pKa = 4.0IMPLYY354 pKa = 10.12YY355 pKa = 10.08QSQDD359 pKa = 3.51VLDD362 pKa = 4.87VIANSTSEE370 pKa = 4.05FMTSRR375 pKa = 11.84DD376 pKa = 3.65FVAKK380 pKa = 10.13AINIGIPLEE389 pKa = 4.14WNLDD393 pKa = 3.57QEE395 pKa = 4.78NLVSQPRR402 pKa = 11.84HH403 pKa = 4.02NVMVKK408 pKa = 10.43SKK410 pKa = 10.74LSPEE414 pKa = 3.91NNIGKK419 pKa = 7.59TLSVNPFTTAWEE431 pKa = 4.11LLTLSFVVDD440 pKa = 3.13WFVNFGDD447 pKa = 4.23VIAGFTGGYY456 pKa = 9.51SDD458 pKa = 6.04DD459 pKa = 4.26SGATASWRR467 pKa = 11.84FDD469 pKa = 3.61DD470 pKa = 5.31KK471 pKa = 11.53KK472 pKa = 11.25VFHH475 pKa = 6.96LKK477 pKa = 10.34NIPSAMVIVDD487 pKa = 2.93INFYY491 pKa = 8.72TRR493 pKa = 11.84QVIDD497 pKa = 3.59PRR499 pKa = 11.84LCGGLAFSPKK509 pKa = 10.36LNLFRR514 pKa = 11.84YY515 pKa = 9.22LDD517 pKa = 3.93AMSLSWNRR525 pKa = 11.84SRR527 pKa = 11.84LKK529 pKa = 10.38ISRR532 pKa = 11.84ATT534 pKa = 3.32
Molecular weight: 60.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1290 |
35 |
590 |
322.5 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.364 ± 1.087 | 1.24 ± 0.64 |
5.736 ± 0.955 | 3.721 ± 0.475 |
5.504 ± 0.935 | 5.271 ± 0.626 |
1.938 ± 0.702 | 6.202 ± 0.248 |
6.202 ± 0.357 | 8.45 ± 1.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.589 | 5.194 ± 0.655 |
4.574 ± 0.513 | 3.488 ± 0.18 |
5.814 ± 1.33 | 8.14 ± 0.718 |
5.969 ± 1.203 | 7.519 ± 0.567 |
2.016 ± 0.355 | 3.333 ± 0.958 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
