
Lake Sarah-associated circular molecule 7
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
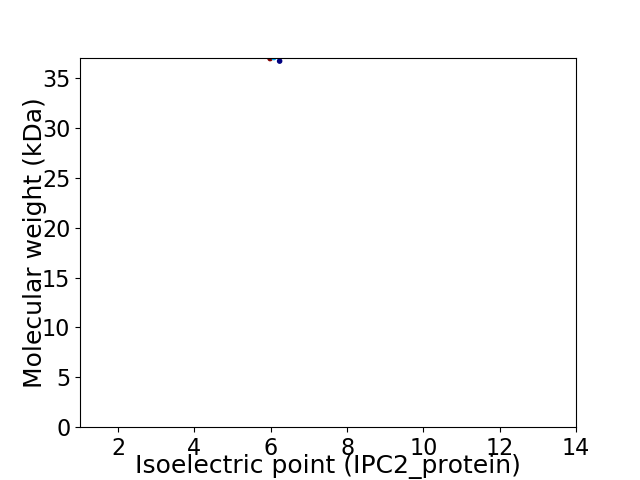
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9G3|A0A126G9G3_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 7 OX=1685732 PE=4 SV=1
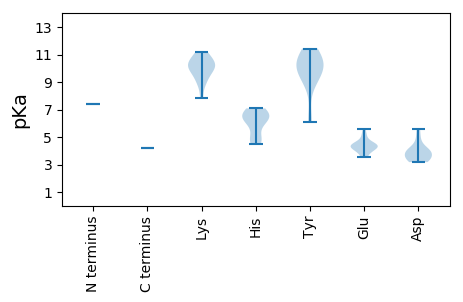
MM1 pKa = 7.43PASSKK6 pKa = 10.24SWCFTDD12 pKa = 4.9NEE14 pKa = 4.43CLEE17 pKa = 4.44DD18 pKa = 3.4LWNIMEE24 pKa = 4.39STYY27 pKa = 11.02LCYY30 pKa = 10.54GRR32 pKa = 11.84EE33 pKa = 4.08TAPTTGQKK41 pKa = 9.97HH42 pKa = 4.56LQGVIVFATQKK53 pKa = 10.54RR54 pKa = 11.84LAAVKK59 pKa = 9.94KK60 pKa = 8.68LHH62 pKa = 6.14PTCHH66 pKa = 6.31WEE68 pKa = 3.96AAHH71 pKa = 6.33SVPASITYY79 pKa = 9.52CKK81 pKa = 10.49KK82 pKa = 9.73EE83 pKa = 3.81GDD85 pKa = 3.36WFEE88 pKa = 5.19FGTPPAQGKK97 pKa = 10.3RR98 pKa = 11.84NDD100 pKa = 3.93LEE102 pKa = 4.32SVYY105 pKa = 11.44ALIKK109 pKa = 9.88TGAKK113 pKa = 9.22FAEE116 pKa = 4.51VLDD119 pKa = 3.95NHH121 pKa = 6.52PVATIHH127 pKa = 5.12YY128 pKa = 6.12TQGIKK133 pKa = 9.74QAIFAQIKK141 pKa = 9.34HH142 pKa = 6.25RR143 pKa = 11.84PEE145 pKa = 3.52QTPTVNWYY153 pKa = 10.3FGATGAGKK161 pKa = 7.82SHH163 pKa = 7.08RR164 pKa = 11.84AHH166 pKa = 6.85AEE168 pKa = 3.54AVNPYY173 pKa = 9.63IFPNTGRR180 pKa = 11.84FWEE183 pKa = 5.57GYY185 pKa = 8.93EE186 pKa = 4.21GQTHH190 pKa = 6.98VIFDD194 pKa = 4.4DD195 pKa = 4.09FRR197 pKa = 11.84PDD199 pKa = 3.21QVPFAKK205 pKa = 10.58LLRR208 pKa = 11.84ILDD211 pKa = 4.03KK212 pKa = 11.19YY213 pKa = 10.48NCTVEE218 pKa = 4.49VKK220 pKa = 10.41GASVPLAATHH230 pKa = 6.38FWITAPEE237 pKa = 4.51SPQEE241 pKa = 4.09MYY243 pKa = 9.81TATNHH248 pKa = 4.94TTGSTWEE255 pKa = 4.38KK256 pKa = 11.09EE257 pKa = 4.2NIQQLVRR264 pKa = 11.84RR265 pKa = 11.84CTVIEE270 pKa = 4.11RR271 pKa = 11.84IIRR274 pKa = 11.84DD275 pKa = 3.37KK276 pKa = 11.11HH277 pKa = 4.48ITNLIIEE284 pKa = 5.0DD285 pKa = 3.72PAGCPEE291 pKa = 4.7RR292 pKa = 11.84GVTDD296 pKa = 3.88IPPLKK301 pKa = 10.1SQKK304 pKa = 10.61KK305 pKa = 9.0LDD307 pKa = 3.86YY308 pKa = 10.45PITDD312 pKa = 3.18HH313 pKa = 6.79MFLFDD318 pKa = 5.55SSDD321 pKa = 3.44EE322 pKa = 4.32FNLCC326 pKa = 4.24
MM1 pKa = 7.43PASSKK6 pKa = 10.24SWCFTDD12 pKa = 4.9NEE14 pKa = 4.43CLEE17 pKa = 4.44DD18 pKa = 3.4LWNIMEE24 pKa = 4.39STYY27 pKa = 11.02LCYY30 pKa = 10.54GRR32 pKa = 11.84EE33 pKa = 4.08TAPTTGQKK41 pKa = 9.97HH42 pKa = 4.56LQGVIVFATQKK53 pKa = 10.54RR54 pKa = 11.84LAAVKK59 pKa = 9.94KK60 pKa = 8.68LHH62 pKa = 6.14PTCHH66 pKa = 6.31WEE68 pKa = 3.96AAHH71 pKa = 6.33SVPASITYY79 pKa = 9.52CKK81 pKa = 10.49KK82 pKa = 9.73EE83 pKa = 3.81GDD85 pKa = 3.36WFEE88 pKa = 5.19FGTPPAQGKK97 pKa = 10.3RR98 pKa = 11.84NDD100 pKa = 3.93LEE102 pKa = 4.32SVYY105 pKa = 11.44ALIKK109 pKa = 9.88TGAKK113 pKa = 9.22FAEE116 pKa = 4.51VLDD119 pKa = 3.95NHH121 pKa = 6.52PVATIHH127 pKa = 5.12YY128 pKa = 6.12TQGIKK133 pKa = 9.74QAIFAQIKK141 pKa = 9.34HH142 pKa = 6.25RR143 pKa = 11.84PEE145 pKa = 3.52QTPTVNWYY153 pKa = 10.3FGATGAGKK161 pKa = 7.82SHH163 pKa = 7.08RR164 pKa = 11.84AHH166 pKa = 6.85AEE168 pKa = 3.54AVNPYY173 pKa = 9.63IFPNTGRR180 pKa = 11.84FWEE183 pKa = 5.57GYY185 pKa = 8.93EE186 pKa = 4.21GQTHH190 pKa = 6.98VIFDD194 pKa = 4.4DD195 pKa = 4.09FRR197 pKa = 11.84PDD199 pKa = 3.21QVPFAKK205 pKa = 10.58LLRR208 pKa = 11.84ILDD211 pKa = 4.03KK212 pKa = 11.19YY213 pKa = 10.48NCTVEE218 pKa = 4.49VKK220 pKa = 10.41GASVPLAATHH230 pKa = 6.38FWITAPEE237 pKa = 4.51SPQEE241 pKa = 4.09MYY243 pKa = 9.81TATNHH248 pKa = 4.94TTGSTWEE255 pKa = 4.38KK256 pKa = 11.09EE257 pKa = 4.2NIQQLVRR264 pKa = 11.84RR265 pKa = 11.84CTVIEE270 pKa = 4.11RR271 pKa = 11.84IIRR274 pKa = 11.84DD275 pKa = 3.37KK276 pKa = 11.11HH277 pKa = 4.48ITNLIIEE284 pKa = 5.0DD285 pKa = 3.72PAGCPEE291 pKa = 4.7RR292 pKa = 11.84GVTDD296 pKa = 3.88IPPLKK301 pKa = 10.1SQKK304 pKa = 10.61KK305 pKa = 9.0LDD307 pKa = 3.86YY308 pKa = 10.45PITDD312 pKa = 3.18HH313 pKa = 6.79MFLFDD318 pKa = 5.55SSDD321 pKa = 3.44EE322 pKa = 4.32FNLCC326 pKa = 4.24
Molecular weight: 36.95 kDa
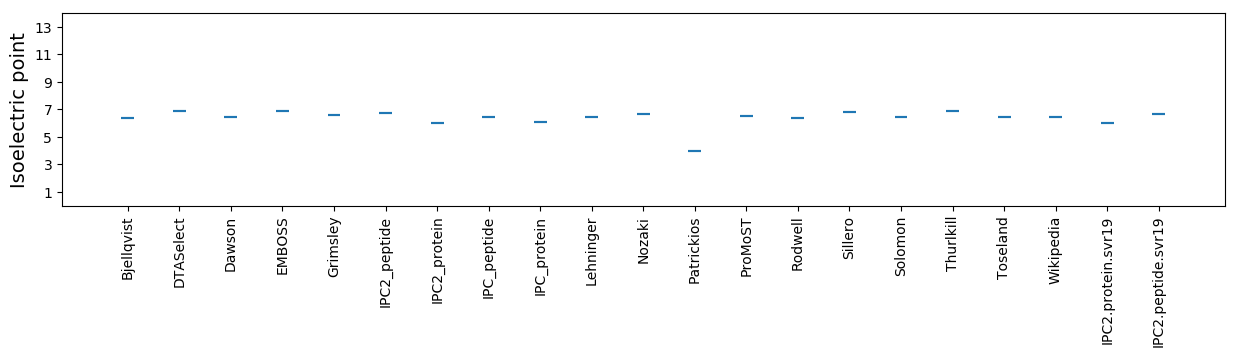
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9G3|A0A126G9G3_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 7 OX=1685732 PE=4 SV=1
MM1 pKa = 7.43PASSKK6 pKa = 10.24SWCFTDD12 pKa = 4.9NEE14 pKa = 4.43CLEE17 pKa = 4.44DD18 pKa = 3.4LWNIMEE24 pKa = 4.39STYY27 pKa = 11.02LCYY30 pKa = 10.54GRR32 pKa = 11.84EE33 pKa = 4.08TAPTTGQKK41 pKa = 9.97HH42 pKa = 4.56LQGVIVFATQKK53 pKa = 10.54RR54 pKa = 11.84LAAVKK59 pKa = 9.94KK60 pKa = 8.68LHH62 pKa = 6.14PTCHH66 pKa = 6.31WEE68 pKa = 3.96AAHH71 pKa = 6.33SVPASITYY79 pKa = 9.52CKK81 pKa = 10.49KK82 pKa = 9.73EE83 pKa = 3.81GDD85 pKa = 3.36WFEE88 pKa = 5.19FGTPPAQGKK97 pKa = 10.3RR98 pKa = 11.84NDD100 pKa = 3.93LEE102 pKa = 4.32SVYY105 pKa = 11.44ALIKK109 pKa = 9.88TGAKK113 pKa = 9.22FAEE116 pKa = 4.51VLDD119 pKa = 3.95NHH121 pKa = 6.52PVATIHH127 pKa = 5.12YY128 pKa = 6.12TQGIKK133 pKa = 9.74QAIFAQIKK141 pKa = 9.34HH142 pKa = 6.25RR143 pKa = 11.84PEE145 pKa = 3.52QTPTVNWYY153 pKa = 10.3FGATGAGKK161 pKa = 7.82SHH163 pKa = 7.08RR164 pKa = 11.84AHH166 pKa = 6.85AEE168 pKa = 3.54AVNPYY173 pKa = 9.63IFPNTGRR180 pKa = 11.84FWEE183 pKa = 5.57GYY185 pKa = 8.93EE186 pKa = 4.21GQTHH190 pKa = 6.98VIFDD194 pKa = 4.4DD195 pKa = 4.09FRR197 pKa = 11.84PDD199 pKa = 3.21QVPFAKK205 pKa = 10.58LLRR208 pKa = 11.84ILDD211 pKa = 4.03KK212 pKa = 11.19YY213 pKa = 10.48NCTVEE218 pKa = 4.49VKK220 pKa = 10.41GASVPLAATHH230 pKa = 6.38FWITAPEE237 pKa = 4.51SPQEE241 pKa = 4.09MYY243 pKa = 9.81TATNHH248 pKa = 4.94TTGSTWEE255 pKa = 4.38KK256 pKa = 11.09EE257 pKa = 4.2NIQQLVRR264 pKa = 11.84RR265 pKa = 11.84CTVIEE270 pKa = 4.11RR271 pKa = 11.84IIRR274 pKa = 11.84DD275 pKa = 3.37KK276 pKa = 11.11HH277 pKa = 4.48ITNLIIEE284 pKa = 5.0DD285 pKa = 3.72PAGCPEE291 pKa = 4.7RR292 pKa = 11.84GVTDD296 pKa = 3.88IPPLKK301 pKa = 10.1SQKK304 pKa = 10.61KK305 pKa = 9.0LDD307 pKa = 3.86YY308 pKa = 10.45PITDD312 pKa = 3.18HH313 pKa = 6.79MFLFDD318 pKa = 5.55SSDD321 pKa = 3.44EE322 pKa = 4.32FNLCC326 pKa = 4.24
MM1 pKa = 7.43PASSKK6 pKa = 10.24SWCFTDD12 pKa = 4.9NEE14 pKa = 4.43CLEE17 pKa = 4.44DD18 pKa = 3.4LWNIMEE24 pKa = 4.39STYY27 pKa = 11.02LCYY30 pKa = 10.54GRR32 pKa = 11.84EE33 pKa = 4.08TAPTTGQKK41 pKa = 9.97HH42 pKa = 4.56LQGVIVFATQKK53 pKa = 10.54RR54 pKa = 11.84LAAVKK59 pKa = 9.94KK60 pKa = 8.68LHH62 pKa = 6.14PTCHH66 pKa = 6.31WEE68 pKa = 3.96AAHH71 pKa = 6.33SVPASITYY79 pKa = 9.52CKK81 pKa = 10.49KK82 pKa = 9.73EE83 pKa = 3.81GDD85 pKa = 3.36WFEE88 pKa = 5.19FGTPPAQGKK97 pKa = 10.3RR98 pKa = 11.84NDD100 pKa = 3.93LEE102 pKa = 4.32SVYY105 pKa = 11.44ALIKK109 pKa = 9.88TGAKK113 pKa = 9.22FAEE116 pKa = 4.51VLDD119 pKa = 3.95NHH121 pKa = 6.52PVATIHH127 pKa = 5.12YY128 pKa = 6.12TQGIKK133 pKa = 9.74QAIFAQIKK141 pKa = 9.34HH142 pKa = 6.25RR143 pKa = 11.84PEE145 pKa = 3.52QTPTVNWYY153 pKa = 10.3FGATGAGKK161 pKa = 7.82SHH163 pKa = 7.08RR164 pKa = 11.84AHH166 pKa = 6.85AEE168 pKa = 3.54AVNPYY173 pKa = 9.63IFPNTGRR180 pKa = 11.84FWEE183 pKa = 5.57GYY185 pKa = 8.93EE186 pKa = 4.21GQTHH190 pKa = 6.98VIFDD194 pKa = 4.4DD195 pKa = 4.09FRR197 pKa = 11.84PDD199 pKa = 3.21QVPFAKK205 pKa = 10.58LLRR208 pKa = 11.84ILDD211 pKa = 4.03KK212 pKa = 11.19YY213 pKa = 10.48NCTVEE218 pKa = 4.49VKK220 pKa = 10.41GASVPLAATHH230 pKa = 6.38FWITAPEE237 pKa = 4.51SPQEE241 pKa = 4.09MYY243 pKa = 9.81TATNHH248 pKa = 4.94TTGSTWEE255 pKa = 4.38KK256 pKa = 11.09EE257 pKa = 4.2NIQQLVRR264 pKa = 11.84RR265 pKa = 11.84CTVIEE270 pKa = 4.11RR271 pKa = 11.84IIRR274 pKa = 11.84DD275 pKa = 3.37KK276 pKa = 11.11HH277 pKa = 4.48ITNLIIEE284 pKa = 5.0DD285 pKa = 3.72PAGCPEE291 pKa = 4.7RR292 pKa = 11.84GVTDD296 pKa = 3.88IPPLKK301 pKa = 10.1SQKK304 pKa = 10.61KK305 pKa = 9.0LDD307 pKa = 3.86YY308 pKa = 10.45PITDD312 pKa = 3.18HH313 pKa = 6.79MFLFDD318 pKa = 5.55SSDD321 pKa = 3.44EE322 pKa = 4.32FNLCC326 pKa = 4.24
Molecular weight: 36.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
326 |
326 |
326 |
326.0 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.282 ± 0.0 | 2.761 ± 0.0 |
4.908 ± 0.0 | 6.748 ± 0.0 |
4.908 ± 0.0 | 5.521 ± 0.0 |
4.294 ± 0.0 | 6.442 ± 0.0 |
6.442 ± 0.0 | 5.828 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.227 ± 0.0 | 3.681 ± 0.0 |
6.442 ± 0.0 | 4.294 ± 0.0 |
3.988 ± 0.0 | 4.294 ± 0.0 |
8.896 ± 0.0 | 5.215 ± 0.0 |
2.454 ± 0.0 | 3.374 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
