
Modestobacter sp. VKM Ac-2676
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Modestobacter; unclassified Modestobacter
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
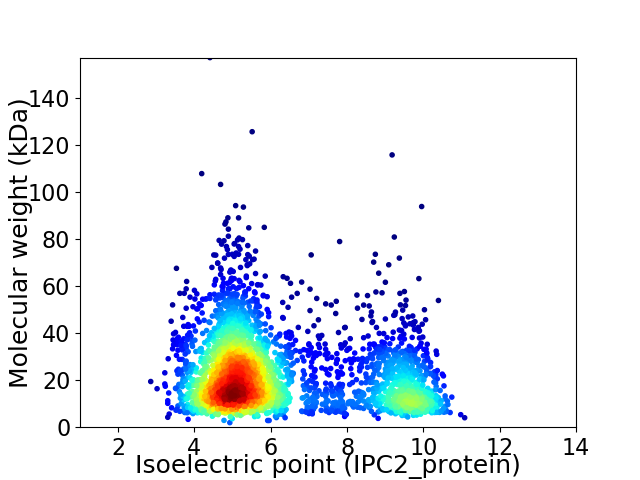
Virtual 2D-PAGE plot for 3187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S8C8D3|A0A1S8C8D3_9ACTN Uncharacterized protein (Fragment) OS=Modestobacter sp. VKM Ac-2676 OX=1678130 GN=A7K94_0212885 PE=4 SV=1
MM1 pKa = 7.29SISWRR6 pKa = 11.84RR7 pKa = 11.84IAGAVLATALGTAALSGCAVTFIEE31 pKa = 5.22GTATPLAGARR41 pKa = 11.84DD42 pKa = 4.11DD43 pKa = 3.88VSPAEE48 pKa = 4.05FPITGAADD56 pKa = 3.69TQVDD60 pKa = 4.98RR61 pKa = 11.84IARR64 pKa = 11.84NALADD69 pKa = 4.45LNTWWAGQFGPVYY82 pKa = 9.59QQQFTPLSGGYY93 pKa = 9.94YY94 pKa = 10.26SVDD97 pKa = 3.51PADD100 pKa = 4.62IDD102 pKa = 3.55PTAYY106 pKa = 9.47PGGEE110 pKa = 4.12IGCGEE115 pKa = 4.2PPEE118 pKa = 4.64AVEE121 pKa = 6.18DD122 pKa = 3.64NAFYY126 pKa = 10.73CGPSQQYY133 pKa = 10.02RR134 pKa = 11.84NSDD137 pKa = 3.94AIQYY141 pKa = 10.28DD142 pKa = 3.82RR143 pKa = 11.84AFLDD147 pKa = 3.45EE148 pKa = 4.53LAFGTGGSEE157 pKa = 4.33GYY159 pKa = 10.84GRR161 pKa = 11.84FIPALVMAHH170 pKa = 6.2EE171 pKa = 5.39FGHH174 pKa = 6.49AVQGRR179 pKa = 11.84VGYY182 pKa = 8.95PFQASIAIEE191 pKa = 4.06TQADD195 pKa = 3.98CFAGAWTRR203 pKa = 11.84WVADD207 pKa = 3.4GEE209 pKa = 4.39APHH212 pKa = 6.6NSIRR216 pKa = 11.84PAEE219 pKa = 4.34LDD221 pKa = 3.5DD222 pKa = 3.8VLRR225 pKa = 11.84GYY227 pKa = 11.18LLLRR231 pKa = 11.84DD232 pKa = 3.93PVGTGLDD239 pKa = 3.13AGEE242 pKa = 4.09AHH244 pKa = 6.97GSYY247 pKa = 10.43FDD249 pKa = 3.76RR250 pKa = 11.84VSAFQEE256 pKa = 4.5GFDD259 pKa = 5.16DD260 pKa = 5.99GPTACRR266 pKa = 11.84DD267 pKa = 3.21GFGPDD272 pKa = 2.81RR273 pKa = 11.84PYY275 pKa = 10.4TQGAFRR281 pKa = 11.84DD282 pKa = 4.03DD283 pKa = 4.56DD284 pKa = 4.32DD285 pKa = 4.93ALSGGDD291 pKa = 3.5APYY294 pKa = 10.33EE295 pKa = 3.88QTVGQFLPEE304 pKa = 3.97GLGEE308 pKa = 4.19FWQQVFAEE316 pKa = 4.27RR317 pKa = 11.84GEE319 pKa = 4.35VFRR322 pKa = 11.84PPTLEE327 pKa = 4.23PFSGRR332 pKa = 11.84APSCDD337 pKa = 3.12GAGADD342 pKa = 4.03VDD344 pKa = 5.54LVYY347 pKa = 11.02CPDD350 pKa = 4.94DD351 pKa = 3.6DD352 pKa = 4.02TVAFDD357 pKa = 3.74EE358 pKa = 4.78AQLTEE363 pKa = 4.14PLYY366 pKa = 10.33EE367 pKa = 4.95AEE369 pKa = 4.41EE370 pKa = 4.84GGDD373 pKa = 3.78YY374 pKa = 11.27AVLTAVAIPYY384 pKa = 9.93GLAGRR389 pKa = 11.84DD390 pKa = 3.57QLGLSTDD397 pKa = 3.76GEE399 pKa = 4.63DD400 pKa = 3.67ALSSAVCLSGAFTASVLNEE419 pKa = 3.86EE420 pKa = 4.49STVLSISPGDD430 pKa = 3.44VDD432 pKa = 3.81EE433 pKa = 5.0SVSFLLEE440 pKa = 4.1YY441 pKa = 9.99ATDD444 pKa = 3.97PQVLPDD450 pKa = 3.62AQLTGFQLVDD460 pKa = 3.08VFRR463 pKa = 11.84TGVFEE468 pKa = 5.08GPAACALDD476 pKa = 3.53RR477 pKa = 4.83
MM1 pKa = 7.29SISWRR6 pKa = 11.84RR7 pKa = 11.84IAGAVLATALGTAALSGCAVTFIEE31 pKa = 5.22GTATPLAGARR41 pKa = 11.84DD42 pKa = 4.11DD43 pKa = 3.88VSPAEE48 pKa = 4.05FPITGAADD56 pKa = 3.69TQVDD60 pKa = 4.98RR61 pKa = 11.84IARR64 pKa = 11.84NALADD69 pKa = 4.45LNTWWAGQFGPVYY82 pKa = 9.59QQQFTPLSGGYY93 pKa = 9.94YY94 pKa = 10.26SVDD97 pKa = 3.51PADD100 pKa = 4.62IDD102 pKa = 3.55PTAYY106 pKa = 9.47PGGEE110 pKa = 4.12IGCGEE115 pKa = 4.2PPEE118 pKa = 4.64AVEE121 pKa = 6.18DD122 pKa = 3.64NAFYY126 pKa = 10.73CGPSQQYY133 pKa = 10.02RR134 pKa = 11.84NSDD137 pKa = 3.94AIQYY141 pKa = 10.28DD142 pKa = 3.82RR143 pKa = 11.84AFLDD147 pKa = 3.45EE148 pKa = 4.53LAFGTGGSEE157 pKa = 4.33GYY159 pKa = 10.84GRR161 pKa = 11.84FIPALVMAHH170 pKa = 6.2EE171 pKa = 5.39FGHH174 pKa = 6.49AVQGRR179 pKa = 11.84VGYY182 pKa = 8.95PFQASIAIEE191 pKa = 4.06TQADD195 pKa = 3.98CFAGAWTRR203 pKa = 11.84WVADD207 pKa = 3.4GEE209 pKa = 4.39APHH212 pKa = 6.6NSIRR216 pKa = 11.84PAEE219 pKa = 4.34LDD221 pKa = 3.5DD222 pKa = 3.8VLRR225 pKa = 11.84GYY227 pKa = 11.18LLLRR231 pKa = 11.84DD232 pKa = 3.93PVGTGLDD239 pKa = 3.13AGEE242 pKa = 4.09AHH244 pKa = 6.97GSYY247 pKa = 10.43FDD249 pKa = 3.76RR250 pKa = 11.84VSAFQEE256 pKa = 4.5GFDD259 pKa = 5.16DD260 pKa = 5.99GPTACRR266 pKa = 11.84DD267 pKa = 3.21GFGPDD272 pKa = 2.81RR273 pKa = 11.84PYY275 pKa = 10.4TQGAFRR281 pKa = 11.84DD282 pKa = 4.03DD283 pKa = 4.56DD284 pKa = 4.32DD285 pKa = 4.93ALSGGDD291 pKa = 3.5APYY294 pKa = 10.33EE295 pKa = 3.88QTVGQFLPEE304 pKa = 3.97GLGEE308 pKa = 4.19FWQQVFAEE316 pKa = 4.27RR317 pKa = 11.84GEE319 pKa = 4.35VFRR322 pKa = 11.84PPTLEE327 pKa = 4.23PFSGRR332 pKa = 11.84APSCDD337 pKa = 3.12GAGADD342 pKa = 4.03VDD344 pKa = 5.54LVYY347 pKa = 11.02CPDD350 pKa = 4.94DD351 pKa = 3.6DD352 pKa = 4.02TVAFDD357 pKa = 3.74EE358 pKa = 4.78AQLTEE363 pKa = 4.14PLYY366 pKa = 10.33EE367 pKa = 4.95AEE369 pKa = 4.41EE370 pKa = 4.84GGDD373 pKa = 3.78YY374 pKa = 11.27AVLTAVAIPYY384 pKa = 9.93GLAGRR389 pKa = 11.84DD390 pKa = 3.57QLGLSTDD397 pKa = 3.76GEE399 pKa = 4.63DD400 pKa = 3.67ALSSAVCLSGAFTASVLNEE419 pKa = 3.86EE420 pKa = 4.49STVLSISPGDD430 pKa = 3.44VDD432 pKa = 3.81EE433 pKa = 5.0SVSFLLEE440 pKa = 4.1YY441 pKa = 9.99ATDD444 pKa = 3.97PQVLPDD450 pKa = 3.62AQLTGFQLVDD460 pKa = 3.08VFRR463 pKa = 11.84TGVFEE468 pKa = 5.08GPAACALDD476 pKa = 3.53RR477 pKa = 4.83
Molecular weight: 50.55 kDa
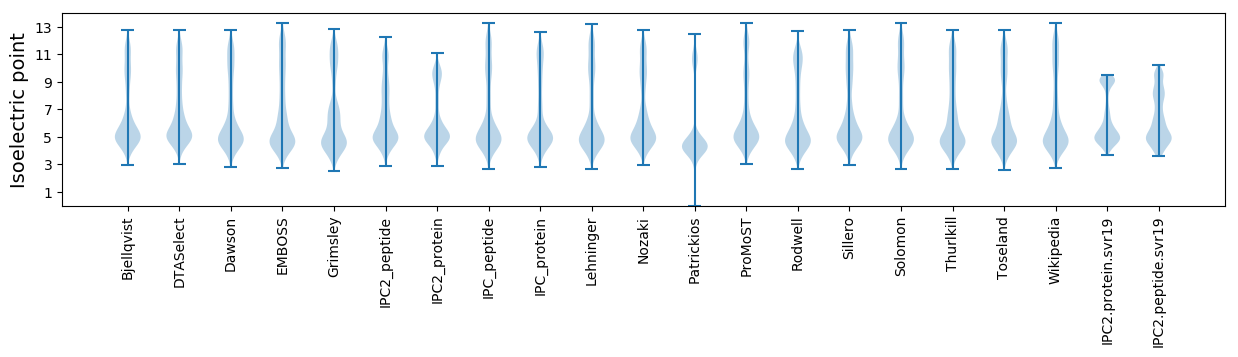
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S8C9V5|A0A1S8C9V5_9ACTN Uncharacterized protein (Fragment) OS=Modestobacter sp. VKM Ac-2676 OX=1678130 GN=A7K94_0207295 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715476 |
18 |
1558 |
224.5 |
23.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.34 ± 0.062 | 0.744 ± 0.014 |
6.189 ± 0.04 | 5.591 ± 0.043 |
2.771 ± 0.026 | 9.603 ± 0.042 |
2.056 ± 0.023 | 3.133 ± 0.034 |
1.438 ± 0.032 | 10.379 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.806 ± 0.017 | 1.586 ± 0.02 |
6.039 ± 0.04 | 2.993 ± 0.028 |
8.022 ± 0.057 | 5.12 ± 0.037 |
6.238 ± 0.036 | 9.625 ± 0.046 |
1.496 ± 0.023 | 1.832 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
