
Coriobacteriaceae bacterium EMTCatB1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; unclassified Coriobacteriaceae
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
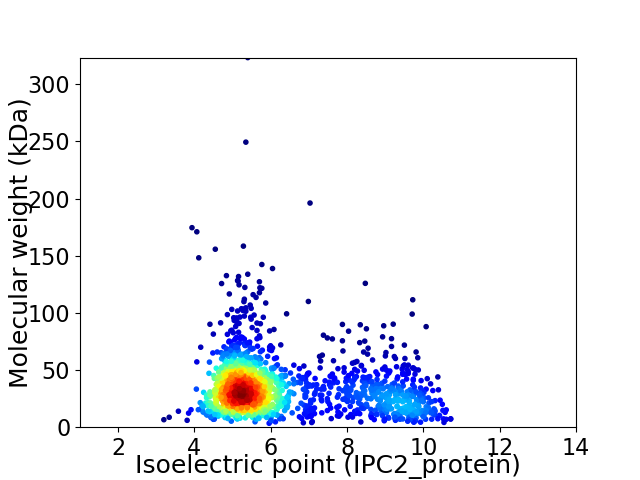
Virtual 2D-PAGE plot for 1659 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1RG50|A0A1V1RG50_9ACTN ATPase related to the helicase subunit of the Holliday junction resolvase OS=Coriobacteriaceae bacterium EMTCatB1 OX=1927122 GN=1c0252 PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84PARR5 pKa = 11.84RR6 pKa = 11.84PLSSKK11 pKa = 10.09EE12 pKa = 3.79AVLRR16 pKa = 11.84GADD19 pKa = 3.26AVYY22 pKa = 10.73DD23 pKa = 3.87GGGATGWFDD32 pKa = 3.43VSPRR36 pKa = 11.84GHH38 pKa = 6.39NSAAAWDD45 pKa = 3.83AGYY48 pKa = 8.73TGAGVKK54 pKa = 9.85IAVADD59 pKa = 4.16DD60 pKa = 4.1SVDD63 pKa = 3.81FAHH66 pKa = 7.82PDD68 pKa = 3.36LQGTQSVVSDD78 pKa = 4.11PASPYY83 pKa = 10.11YY84 pKa = 10.36GWPEE88 pKa = 3.74AFDD91 pKa = 4.51PYY93 pKa = 11.18SALLYY98 pKa = 10.59AYY100 pKa = 9.39DD101 pKa = 3.62VYY103 pKa = 11.64YY104 pKa = 9.19GTSYY108 pKa = 10.25VANGQTWWSDD118 pKa = 2.81TSATITEE125 pKa = 4.65ADD127 pKa = 3.51PTFDD131 pKa = 2.84GKK133 pKa = 9.09TFVTPGTSLSGTYY146 pKa = 10.03HH147 pKa = 7.22IGYY150 pKa = 9.38LWDD153 pKa = 3.64EE154 pKa = 4.3NLYY157 pKa = 11.29AWMSDD162 pKa = 2.15WSEE165 pKa = 4.19YY166 pKa = 8.66PTVLVADD173 pKa = 3.87EE174 pKa = 4.45TTPGVYY180 pKa = 9.07DD181 pKa = 3.4TVYY184 pKa = 10.32VDD186 pKa = 2.63VWMDD190 pKa = 3.24YY191 pKa = 11.13DD192 pKa = 4.42FSYY195 pKa = 10.95EE196 pKa = 4.16KK197 pKa = 10.32PCTIDD202 pKa = 3.97SPISYY207 pKa = 10.36LDD209 pKa = 3.47YY210 pKa = 10.37WDD212 pKa = 5.52SEE214 pKa = 4.28ADD216 pKa = 3.62DD217 pKa = 4.57YY218 pKa = 12.18GPDD221 pKa = 3.61GYY223 pKa = 11.63ADD225 pKa = 3.81LSGGTVYY232 pKa = 10.42WISDD236 pKa = 3.97GEE238 pKa = 4.2HH239 pKa = 5.65QPPGFEE245 pKa = 4.04FLINGSDD252 pKa = 3.56PSAPVPGNGEE262 pKa = 4.19LVCFMGALNFDD273 pKa = 4.37EE274 pKa = 4.87NHH276 pKa = 5.82GTLCASNVVGQGMTDD291 pKa = 3.67GPSDD295 pKa = 4.79PIMDD299 pKa = 4.87DD300 pKa = 3.18GVYY303 pKa = 10.4PPFKK307 pKa = 9.91TPTGGGDD314 pKa = 3.87GIVQGAARR322 pKa = 11.84DD323 pKa = 3.95SKK325 pKa = 11.31VVAFSDD331 pKa = 3.64IYY333 pKa = 10.8WNFFTSTLLAYY344 pKa = 10.16DD345 pKa = 3.96YY346 pKa = 10.39ATFGADD352 pKa = 3.29MEE354 pKa = 5.33AGTGDD359 pKa = 4.05EE360 pKa = 4.12IQIVSNSYY368 pKa = 10.77GDD370 pKa = 3.8SAVDD374 pKa = 3.3ADD376 pKa = 3.3EE377 pKa = 4.11WDD379 pKa = 3.47YY380 pKa = 11.62HH381 pKa = 5.95SRR383 pKa = 11.84YY384 pKa = 8.12VTALNTYY391 pKa = 8.14VNSMTSFLFSTGNGAPGYY409 pKa = 7.73GTNAPPTPSTGIGIGASTQLGACGGWDD436 pKa = 3.62SIYY439 pKa = 10.98DD440 pKa = 3.78ADD442 pKa = 3.88QVTVGDD448 pKa = 4.02VIPWSNRR455 pKa = 11.84GPSAMGHH462 pKa = 6.81AGPAVVADD470 pKa = 4.31GAYY473 pKa = 10.52SSGAMALNQGAWDD486 pKa = 4.14GWRR489 pKa = 11.84SWIVWGGTSRR499 pKa = 11.84SCPVAAGNMALIYY512 pKa = 10.5DD513 pKa = 4.17AFKK516 pKa = 10.94QRR518 pKa = 11.84FGRR521 pKa = 11.84YY522 pKa = 4.06PTYY525 pKa = 9.2WQARR529 pKa = 11.84NYY531 pKa = 10.7LMAGARR537 pKa = 11.84DD538 pKa = 3.99LSYY541 pKa = 11.52DD542 pKa = 3.69PAVQGAGMVDD552 pKa = 3.16AKK554 pKa = 10.85KK555 pKa = 10.84SIDD558 pKa = 3.86IITGDD563 pKa = 3.59GGIEE567 pKa = 3.7ISPSQWYY574 pKa = 8.45PGDD577 pKa = 3.79YY578 pKa = 10.18RR579 pKa = 11.84GEE581 pKa = 4.36TYY583 pKa = 11.02ASFPSVVYY591 pKa = 10.12PGQTYY596 pKa = 10.81DD597 pKa = 3.55GTLQVDD603 pKa = 3.54NRR605 pKa = 11.84STTDD609 pKa = 2.56SDD611 pKa = 4.43VNVTVSDD618 pKa = 3.65KK619 pKa = 10.77TLEE622 pKa = 4.14PVAASSMLVEE632 pKa = 4.53LDD634 pKa = 3.4GSKK637 pKa = 10.57EE638 pKa = 3.95SPYY641 pKa = 11.3DD642 pKa = 3.48FNRR645 pKa = 11.84PDD647 pKa = 3.28WLSDD651 pKa = 3.18LSALVRR657 pKa = 11.84DD658 pKa = 4.18YY659 pKa = 11.52DD660 pKa = 3.96PDD662 pKa = 3.88LMVVRR667 pKa = 11.84TATPFADD674 pKa = 4.92FAPTGAFNTSGSTHH688 pKa = 5.12NTTRR692 pKa = 11.84LLAYY696 pKa = 9.65DD697 pKa = 4.17WKK699 pKa = 11.1DD700 pKa = 3.29QDD702 pKa = 3.69EE703 pKa = 5.83DD704 pKa = 4.08GVLWNDD710 pKa = 3.23GDD712 pKa = 3.91GDD714 pKa = 4.49GYY716 pKa = 11.33VDD718 pKa = 4.33PGEE721 pKa = 4.29MDD723 pKa = 2.73SGEE726 pKa = 4.19YY727 pKa = 9.64MRR729 pKa = 11.84FTYY732 pKa = 10.84SNNFANSHH740 pKa = 6.44EE741 pKa = 4.13IRR743 pKa = 11.84VQNPVEE749 pKa = 4.05RR750 pKa = 11.84MHH752 pKa = 7.56DD753 pKa = 4.08GIFLGLQHH761 pKa = 6.7NNDD764 pKa = 3.03ATADD768 pKa = 3.7KK769 pKa = 9.29TVSVQIVVEE778 pKa = 4.55LYY780 pKa = 10.73KK781 pKa = 10.53EE782 pKa = 4.46TDD784 pKa = 3.71HH785 pKa = 6.96PWLSSSMEE793 pKa = 4.04GSPHH797 pKa = 7.78LLMGGSSMQVPVRR810 pKa = 11.84LTVPEE815 pKa = 4.12DD816 pKa = 3.51AKK818 pKa = 10.66PGVYY822 pKa = 9.83EE823 pKa = 4.34GEE825 pKa = 4.36FVFTTDD831 pKa = 4.47RR832 pKa = 11.84GGDD835 pKa = 3.52LYY837 pKa = 10.9TSVVPVHH844 pKa = 5.9ITVASDD850 pKa = 3.51SPNFAFGDD858 pKa = 3.58VGMVEE863 pKa = 4.17PRR865 pKa = 11.84GLRR868 pKa = 11.84PTGLGPADD876 pKa = 4.62PSPLLPNYY884 pKa = 9.83QLFGGQSWNWRR895 pKa = 11.84AEE897 pKa = 4.05SGDD900 pKa = 3.0WRR902 pKa = 11.84FYY904 pKa = 10.51FADD907 pKa = 3.85VKK909 pKa = 11.22DD910 pKa = 4.47EE911 pKa = 4.21EE912 pKa = 4.68PLPAGATWLVKK923 pKa = 7.34TTWPDD928 pKa = 2.57SGQPVDD934 pKa = 3.8TDD936 pKa = 3.22IDD938 pKa = 3.83TLLYY942 pKa = 11.0GPVDD946 pKa = 4.71DD947 pKa = 5.5GWSSMDD953 pKa = 3.48PGVFGPHH960 pKa = 5.31GMEE963 pKa = 4.2LKK965 pKa = 10.73GGSSNTNIGSGIWLWQTTTGTTEE988 pKa = 3.64EE989 pKa = 4.51WVAGPLEE996 pKa = 4.58RR997 pKa = 11.84GLNEE1001 pKa = 3.75IMLHH1005 pKa = 4.72SVVWSGEE1012 pKa = 3.82SWRR1015 pKa = 11.84EE1016 pKa = 3.86PVSGEE1021 pKa = 3.87AGVVGVDD1028 pKa = 3.46PSSIDD1033 pKa = 3.31VCDD1036 pKa = 3.68SAGSGSVPLQFTTTMDD1052 pKa = 3.98LNGLASEE1059 pKa = 4.81SYY1061 pKa = 9.42GVTRR1065 pKa = 11.84RR1066 pKa = 11.84LHH1068 pKa = 5.89YY1069 pKa = 9.39PAEE1072 pKa = 4.69SISQDD1077 pKa = 2.64EE1078 pKa = 4.18DD1079 pKa = 3.98VYY1081 pKa = 11.83YY1082 pKa = 10.86DD1083 pKa = 4.36LEE1085 pKa = 4.88LSGAAYY1091 pKa = 10.78LDD1093 pKa = 3.75VSISAAGGSDD1103 pKa = 3.43LDD1105 pKa = 5.15LYY1107 pKa = 9.13VWWWDD1112 pKa = 3.32GSSWQLVGASEE1123 pKa = 4.1TATGDD1128 pKa = 3.02EE1129 pKa = 4.91HH1130 pKa = 8.99VRR1132 pKa = 11.84IDD1134 pKa = 3.55QPADD1138 pKa = 2.82GSYY1141 pKa = 10.56VVDD1144 pKa = 3.88VYY1146 pKa = 11.24GYY1148 pKa = 9.88SVSGTDD1154 pKa = 3.07TFEE1157 pKa = 3.82ITASYY1162 pKa = 10.62PMGDD1166 pKa = 3.73DD1167 pKa = 3.41VTVTGLPAGPLVAGQVVPMSVDD1189 pKa = 2.69WTKK1192 pKa = 11.0YY1193 pKa = 10.31RR1194 pKa = 11.84GDD1196 pKa = 3.55LPDD1199 pKa = 4.08RR1200 pKa = 11.84EE1201 pKa = 4.47GVYY1204 pKa = 10.52EE1205 pKa = 4.08GVVYY1209 pKa = 10.58LGPTEE1214 pKa = 4.19APRR1217 pKa = 11.84AIAIPFEE1224 pKa = 3.84LSYY1227 pKa = 10.71PFEE1230 pKa = 4.47VEE1232 pKa = 4.0QYY1234 pKa = 9.54TPNPSQPAMSPTTAIEE1250 pKa = 3.88IQFSKK1255 pKa = 10.88RR1256 pKa = 11.84LDD1258 pKa = 3.54EE1259 pKa = 5.53DD1260 pKa = 4.13SIGDD1264 pKa = 3.7GTFTLTDD1271 pKa = 3.55GTEE1274 pKa = 4.23EE1275 pKa = 4.01IPLDD1279 pKa = 3.7VSADD1283 pKa = 3.44AEE1285 pKa = 4.47TGHH1288 pKa = 6.79VVLTPQDD1295 pKa = 3.6MLKK1298 pKa = 10.65SSTEE1302 pKa = 3.79YY1303 pKa = 10.7LVEE1306 pKa = 4.68IDD1308 pKa = 4.59GLLSRR1313 pKa = 11.84DD1314 pKa = 3.26GDD1316 pKa = 4.12EE1317 pKa = 5.63LSTALPFSTTDD1328 pKa = 3.66AIVRR1332 pKa = 11.84AMGDD1336 pKa = 3.23TRR1338 pKa = 11.84YY1339 pKa = 8.79DD1340 pKa = 3.24TAVAVSQQSFDD1351 pKa = 3.36AAEE1354 pKa = 4.15AVVLATGEE1362 pKa = 4.28KK1363 pKa = 10.63FPDD1366 pKa = 3.55ALSASGLAGQVKK1378 pKa = 10.44GPVLLTKK1385 pKa = 10.09PDD1387 pKa = 3.5TLLPQVRR1394 pKa = 11.84DD1395 pKa = 3.85EE1396 pKa = 4.1IARR1399 pKa = 11.84LGATKK1404 pKa = 10.37VYY1406 pKa = 10.7LIGGTGALSSNVASAVDD1423 pKa = 3.66ALPGVSIEE1431 pKa = 4.25RR1432 pKa = 11.84VQGADD1437 pKa = 3.15RR1438 pKa = 11.84YY1439 pKa = 8.89ATAAEE1444 pKa = 4.34VARR1447 pKa = 11.84KK1448 pKa = 9.02IAALRR1453 pKa = 11.84GSALSDD1459 pKa = 3.18AFLVRR1464 pKa = 11.84GDD1466 pKa = 3.96EE1467 pKa = 4.31YY1468 pKa = 11.53ADD1470 pKa = 3.59GVAVSPYY1477 pKa = 10.31AYY1479 pKa = 9.32RR1480 pKa = 11.84QAMPVLLTMPNDD1492 pKa = 3.54LPAVTRR1498 pKa = 11.84SALSDD1503 pKa = 3.59LSVSDD1508 pKa = 3.09VWIAGGVGAVSNRR1521 pKa = 11.84VVGQLPSGTTSHH1533 pKa = 6.83RR1534 pKa = 11.84MGGANRR1540 pKa = 11.84YY1541 pKa = 7.29ATAATIAQTAVGNSWAVWTVVGVATGRR1568 pKa = 11.84NFPDD1572 pKa = 3.71ALTGGVGIGKK1582 pKa = 9.39QKK1584 pKa = 11.01GVMLLTEE1591 pKa = 4.29PTVLSPEE1598 pKa = 4.06TKK1600 pKa = 9.55TAIEE1604 pKa = 4.39SNASEE1609 pKa = 4.05ILKK1612 pKa = 10.45LSVIGGPGAVSAAVSNAIADD1632 pKa = 4.36LLWW1635 pKa = 5.21
MM1 pKa = 7.58RR2 pKa = 11.84PARR5 pKa = 11.84RR6 pKa = 11.84PLSSKK11 pKa = 10.09EE12 pKa = 3.79AVLRR16 pKa = 11.84GADD19 pKa = 3.26AVYY22 pKa = 10.73DD23 pKa = 3.87GGGATGWFDD32 pKa = 3.43VSPRR36 pKa = 11.84GHH38 pKa = 6.39NSAAAWDD45 pKa = 3.83AGYY48 pKa = 8.73TGAGVKK54 pKa = 9.85IAVADD59 pKa = 4.16DD60 pKa = 4.1SVDD63 pKa = 3.81FAHH66 pKa = 7.82PDD68 pKa = 3.36LQGTQSVVSDD78 pKa = 4.11PASPYY83 pKa = 10.11YY84 pKa = 10.36GWPEE88 pKa = 3.74AFDD91 pKa = 4.51PYY93 pKa = 11.18SALLYY98 pKa = 10.59AYY100 pKa = 9.39DD101 pKa = 3.62VYY103 pKa = 11.64YY104 pKa = 9.19GTSYY108 pKa = 10.25VANGQTWWSDD118 pKa = 2.81TSATITEE125 pKa = 4.65ADD127 pKa = 3.51PTFDD131 pKa = 2.84GKK133 pKa = 9.09TFVTPGTSLSGTYY146 pKa = 10.03HH147 pKa = 7.22IGYY150 pKa = 9.38LWDD153 pKa = 3.64EE154 pKa = 4.3NLYY157 pKa = 11.29AWMSDD162 pKa = 2.15WSEE165 pKa = 4.19YY166 pKa = 8.66PTVLVADD173 pKa = 3.87EE174 pKa = 4.45TTPGVYY180 pKa = 9.07DD181 pKa = 3.4TVYY184 pKa = 10.32VDD186 pKa = 2.63VWMDD190 pKa = 3.24YY191 pKa = 11.13DD192 pKa = 4.42FSYY195 pKa = 10.95EE196 pKa = 4.16KK197 pKa = 10.32PCTIDD202 pKa = 3.97SPISYY207 pKa = 10.36LDD209 pKa = 3.47YY210 pKa = 10.37WDD212 pKa = 5.52SEE214 pKa = 4.28ADD216 pKa = 3.62DD217 pKa = 4.57YY218 pKa = 12.18GPDD221 pKa = 3.61GYY223 pKa = 11.63ADD225 pKa = 3.81LSGGTVYY232 pKa = 10.42WISDD236 pKa = 3.97GEE238 pKa = 4.2HH239 pKa = 5.65QPPGFEE245 pKa = 4.04FLINGSDD252 pKa = 3.56PSAPVPGNGEE262 pKa = 4.19LVCFMGALNFDD273 pKa = 4.37EE274 pKa = 4.87NHH276 pKa = 5.82GTLCASNVVGQGMTDD291 pKa = 3.67GPSDD295 pKa = 4.79PIMDD299 pKa = 4.87DD300 pKa = 3.18GVYY303 pKa = 10.4PPFKK307 pKa = 9.91TPTGGGDD314 pKa = 3.87GIVQGAARR322 pKa = 11.84DD323 pKa = 3.95SKK325 pKa = 11.31VVAFSDD331 pKa = 3.64IYY333 pKa = 10.8WNFFTSTLLAYY344 pKa = 10.16DD345 pKa = 3.96YY346 pKa = 10.39ATFGADD352 pKa = 3.29MEE354 pKa = 5.33AGTGDD359 pKa = 4.05EE360 pKa = 4.12IQIVSNSYY368 pKa = 10.77GDD370 pKa = 3.8SAVDD374 pKa = 3.3ADD376 pKa = 3.3EE377 pKa = 4.11WDD379 pKa = 3.47YY380 pKa = 11.62HH381 pKa = 5.95SRR383 pKa = 11.84YY384 pKa = 8.12VTALNTYY391 pKa = 8.14VNSMTSFLFSTGNGAPGYY409 pKa = 7.73GTNAPPTPSTGIGIGASTQLGACGGWDD436 pKa = 3.62SIYY439 pKa = 10.98DD440 pKa = 3.78ADD442 pKa = 3.88QVTVGDD448 pKa = 4.02VIPWSNRR455 pKa = 11.84GPSAMGHH462 pKa = 6.81AGPAVVADD470 pKa = 4.31GAYY473 pKa = 10.52SSGAMALNQGAWDD486 pKa = 4.14GWRR489 pKa = 11.84SWIVWGGTSRR499 pKa = 11.84SCPVAAGNMALIYY512 pKa = 10.5DD513 pKa = 4.17AFKK516 pKa = 10.94QRR518 pKa = 11.84FGRR521 pKa = 11.84YY522 pKa = 4.06PTYY525 pKa = 9.2WQARR529 pKa = 11.84NYY531 pKa = 10.7LMAGARR537 pKa = 11.84DD538 pKa = 3.99LSYY541 pKa = 11.52DD542 pKa = 3.69PAVQGAGMVDD552 pKa = 3.16AKK554 pKa = 10.85KK555 pKa = 10.84SIDD558 pKa = 3.86IITGDD563 pKa = 3.59GGIEE567 pKa = 3.7ISPSQWYY574 pKa = 8.45PGDD577 pKa = 3.79YY578 pKa = 10.18RR579 pKa = 11.84GEE581 pKa = 4.36TYY583 pKa = 11.02ASFPSVVYY591 pKa = 10.12PGQTYY596 pKa = 10.81DD597 pKa = 3.55GTLQVDD603 pKa = 3.54NRR605 pKa = 11.84STTDD609 pKa = 2.56SDD611 pKa = 4.43VNVTVSDD618 pKa = 3.65KK619 pKa = 10.77TLEE622 pKa = 4.14PVAASSMLVEE632 pKa = 4.53LDD634 pKa = 3.4GSKK637 pKa = 10.57EE638 pKa = 3.95SPYY641 pKa = 11.3DD642 pKa = 3.48FNRR645 pKa = 11.84PDD647 pKa = 3.28WLSDD651 pKa = 3.18LSALVRR657 pKa = 11.84DD658 pKa = 4.18YY659 pKa = 11.52DD660 pKa = 3.96PDD662 pKa = 3.88LMVVRR667 pKa = 11.84TATPFADD674 pKa = 4.92FAPTGAFNTSGSTHH688 pKa = 5.12NTTRR692 pKa = 11.84LLAYY696 pKa = 9.65DD697 pKa = 4.17WKK699 pKa = 11.1DD700 pKa = 3.29QDD702 pKa = 3.69EE703 pKa = 5.83DD704 pKa = 4.08GVLWNDD710 pKa = 3.23GDD712 pKa = 3.91GDD714 pKa = 4.49GYY716 pKa = 11.33VDD718 pKa = 4.33PGEE721 pKa = 4.29MDD723 pKa = 2.73SGEE726 pKa = 4.19YY727 pKa = 9.64MRR729 pKa = 11.84FTYY732 pKa = 10.84SNNFANSHH740 pKa = 6.44EE741 pKa = 4.13IRR743 pKa = 11.84VQNPVEE749 pKa = 4.05RR750 pKa = 11.84MHH752 pKa = 7.56DD753 pKa = 4.08GIFLGLQHH761 pKa = 6.7NNDD764 pKa = 3.03ATADD768 pKa = 3.7KK769 pKa = 9.29TVSVQIVVEE778 pKa = 4.55LYY780 pKa = 10.73KK781 pKa = 10.53EE782 pKa = 4.46TDD784 pKa = 3.71HH785 pKa = 6.96PWLSSSMEE793 pKa = 4.04GSPHH797 pKa = 7.78LLMGGSSMQVPVRR810 pKa = 11.84LTVPEE815 pKa = 4.12DD816 pKa = 3.51AKK818 pKa = 10.66PGVYY822 pKa = 9.83EE823 pKa = 4.34GEE825 pKa = 4.36FVFTTDD831 pKa = 4.47RR832 pKa = 11.84GGDD835 pKa = 3.52LYY837 pKa = 10.9TSVVPVHH844 pKa = 5.9ITVASDD850 pKa = 3.51SPNFAFGDD858 pKa = 3.58VGMVEE863 pKa = 4.17PRR865 pKa = 11.84GLRR868 pKa = 11.84PTGLGPADD876 pKa = 4.62PSPLLPNYY884 pKa = 9.83QLFGGQSWNWRR895 pKa = 11.84AEE897 pKa = 4.05SGDD900 pKa = 3.0WRR902 pKa = 11.84FYY904 pKa = 10.51FADD907 pKa = 3.85VKK909 pKa = 11.22DD910 pKa = 4.47EE911 pKa = 4.21EE912 pKa = 4.68PLPAGATWLVKK923 pKa = 7.34TTWPDD928 pKa = 2.57SGQPVDD934 pKa = 3.8TDD936 pKa = 3.22IDD938 pKa = 3.83TLLYY942 pKa = 11.0GPVDD946 pKa = 4.71DD947 pKa = 5.5GWSSMDD953 pKa = 3.48PGVFGPHH960 pKa = 5.31GMEE963 pKa = 4.2LKK965 pKa = 10.73GGSSNTNIGSGIWLWQTTTGTTEE988 pKa = 3.64EE989 pKa = 4.51WVAGPLEE996 pKa = 4.58RR997 pKa = 11.84GLNEE1001 pKa = 3.75IMLHH1005 pKa = 4.72SVVWSGEE1012 pKa = 3.82SWRR1015 pKa = 11.84EE1016 pKa = 3.86PVSGEE1021 pKa = 3.87AGVVGVDD1028 pKa = 3.46PSSIDD1033 pKa = 3.31VCDD1036 pKa = 3.68SAGSGSVPLQFTTTMDD1052 pKa = 3.98LNGLASEE1059 pKa = 4.81SYY1061 pKa = 9.42GVTRR1065 pKa = 11.84RR1066 pKa = 11.84LHH1068 pKa = 5.89YY1069 pKa = 9.39PAEE1072 pKa = 4.69SISQDD1077 pKa = 2.64EE1078 pKa = 4.18DD1079 pKa = 3.98VYY1081 pKa = 11.83YY1082 pKa = 10.86DD1083 pKa = 4.36LEE1085 pKa = 4.88LSGAAYY1091 pKa = 10.78LDD1093 pKa = 3.75VSISAAGGSDD1103 pKa = 3.43LDD1105 pKa = 5.15LYY1107 pKa = 9.13VWWWDD1112 pKa = 3.32GSSWQLVGASEE1123 pKa = 4.1TATGDD1128 pKa = 3.02EE1129 pKa = 4.91HH1130 pKa = 8.99VRR1132 pKa = 11.84IDD1134 pKa = 3.55QPADD1138 pKa = 2.82GSYY1141 pKa = 10.56VVDD1144 pKa = 3.88VYY1146 pKa = 11.24GYY1148 pKa = 9.88SVSGTDD1154 pKa = 3.07TFEE1157 pKa = 3.82ITASYY1162 pKa = 10.62PMGDD1166 pKa = 3.73DD1167 pKa = 3.41VTVTGLPAGPLVAGQVVPMSVDD1189 pKa = 2.69WTKK1192 pKa = 11.0YY1193 pKa = 10.31RR1194 pKa = 11.84GDD1196 pKa = 3.55LPDD1199 pKa = 4.08RR1200 pKa = 11.84EE1201 pKa = 4.47GVYY1204 pKa = 10.52EE1205 pKa = 4.08GVVYY1209 pKa = 10.58LGPTEE1214 pKa = 4.19APRR1217 pKa = 11.84AIAIPFEE1224 pKa = 3.84LSYY1227 pKa = 10.71PFEE1230 pKa = 4.47VEE1232 pKa = 4.0QYY1234 pKa = 9.54TPNPSQPAMSPTTAIEE1250 pKa = 3.88IQFSKK1255 pKa = 10.88RR1256 pKa = 11.84LDD1258 pKa = 3.54EE1259 pKa = 5.53DD1260 pKa = 4.13SIGDD1264 pKa = 3.7GTFTLTDD1271 pKa = 3.55GTEE1274 pKa = 4.23EE1275 pKa = 4.01IPLDD1279 pKa = 3.7VSADD1283 pKa = 3.44AEE1285 pKa = 4.47TGHH1288 pKa = 6.79VVLTPQDD1295 pKa = 3.6MLKK1298 pKa = 10.65SSTEE1302 pKa = 3.79YY1303 pKa = 10.7LVEE1306 pKa = 4.68IDD1308 pKa = 4.59GLLSRR1313 pKa = 11.84DD1314 pKa = 3.26GDD1316 pKa = 4.12EE1317 pKa = 5.63LSTALPFSTTDD1328 pKa = 3.66AIVRR1332 pKa = 11.84AMGDD1336 pKa = 3.23TRR1338 pKa = 11.84YY1339 pKa = 8.79DD1340 pKa = 3.24TAVAVSQQSFDD1351 pKa = 3.36AAEE1354 pKa = 4.15AVVLATGEE1362 pKa = 4.28KK1363 pKa = 10.63FPDD1366 pKa = 3.55ALSASGLAGQVKK1378 pKa = 10.44GPVLLTKK1385 pKa = 10.09PDD1387 pKa = 3.5TLLPQVRR1394 pKa = 11.84DD1395 pKa = 3.85EE1396 pKa = 4.1IARR1399 pKa = 11.84LGATKK1404 pKa = 10.37VYY1406 pKa = 10.7LIGGTGALSSNVASAVDD1423 pKa = 3.66ALPGVSIEE1431 pKa = 4.25RR1432 pKa = 11.84VQGADD1437 pKa = 3.15RR1438 pKa = 11.84YY1439 pKa = 8.89ATAAEE1444 pKa = 4.34VARR1447 pKa = 11.84KK1448 pKa = 9.02IAALRR1453 pKa = 11.84GSALSDD1459 pKa = 3.18AFLVRR1464 pKa = 11.84GDD1466 pKa = 3.96EE1467 pKa = 4.31YY1468 pKa = 11.53ADD1470 pKa = 3.59GVAVSPYY1477 pKa = 10.31AYY1479 pKa = 9.32RR1480 pKa = 11.84QAMPVLLTMPNDD1492 pKa = 3.54LPAVTRR1498 pKa = 11.84SALSDD1503 pKa = 3.59LSVSDD1508 pKa = 3.09VWIAGGVGAVSNRR1521 pKa = 11.84VVGQLPSGTTSHH1533 pKa = 6.83RR1534 pKa = 11.84MGGANRR1540 pKa = 11.84YY1541 pKa = 7.29ATAATIAQTAVGNSWAVWTVVGVATGRR1568 pKa = 11.84NFPDD1572 pKa = 3.71ALTGGVGIGKK1582 pKa = 9.39QKK1584 pKa = 11.01GVMLLTEE1591 pKa = 4.29PTVLSPEE1598 pKa = 4.06TKK1600 pKa = 9.55TAIEE1604 pKa = 4.39SNASEE1609 pKa = 4.05ILKK1612 pKa = 10.45LSVIGGPGAVSAAVSNAIADD1632 pKa = 4.36LLWW1635 pKa = 5.21
Molecular weight: 174.51 kDa
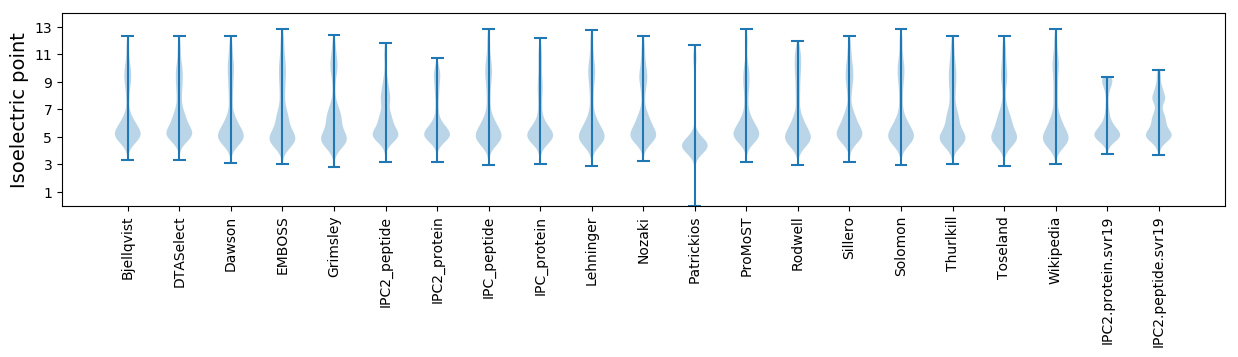
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1REZ6|A0A1V1REZ6_9ACTN NDH-1 subunit N 1 OS=Coriobacteriaceae bacterium EMTCatB1 OX=1927122 GN=nuoN PE=4 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84AVLAGVVRR10 pKa = 11.84RR11 pKa = 11.84AQPARR16 pKa = 11.84HH17 pKa = 5.72HH18 pKa = 6.37RR19 pKa = 11.84GARR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84DD26 pKa = 3.18HH27 pKa = 6.62AGSRR31 pKa = 11.84KK32 pKa = 9.67RR33 pKa = 11.84PSCVAGRR40 pKa = 11.84CVRR43 pKa = 11.84RR44 pKa = 11.84VVWQRR49 pKa = 11.84AGDD52 pKa = 3.74RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.83GVAAGGSRR63 pKa = 11.84SCGGAVAPAFALPDD77 pKa = 3.43LRR79 pKa = 11.84QEE81 pKa = 3.81PRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84AEE87 pKa = 3.62RR88 pKa = 11.84GEE90 pKa = 4.04RR91 pKa = 11.84LMGALLVSIGLPLAITVVIVAGGARR116 pKa = 11.84SVRR119 pKa = 11.84VTRR122 pKa = 11.84VFALAGPLAALAAGASLIASAHH144 pKa = 5.92GGAAAPSWNGAWLAQGGAAAPVGFAGDD171 pKa = 3.46ALAALMLVIVGAVAACVVAFSWGYY195 pKa = 8.18MAHH198 pKa = 7.38EE199 pKa = 5.39DD200 pKa = 3.85GQPRR204 pKa = 11.84YY205 pKa = 9.73YY206 pKa = 10.75AVLMLFVSAMSVLVLADD223 pKa = 3.5NLVTLFIGWEE233 pKa = 3.97LVGACSYY240 pKa = 11.33LLIGFWYY247 pKa = 9.69RR248 pKa = 11.84KK249 pKa = 8.5PAATRR254 pKa = 11.84AAIKK258 pKa = 10.7AFMTTRR264 pKa = 11.84VGDD267 pKa = 3.54VAMLVGMAVLWRR279 pKa = 11.84HH280 pKa = 6.03AGTLSVRR287 pKa = 11.84EE288 pKa = 3.95IVAALPRR295 pKa = 11.84MEE297 pKa = 5.44PGVITAAALLLFGGAAGKK315 pKa = 9.66SAQFPLHH322 pKa = 5.89IWLPDD327 pKa = 3.24AMEE330 pKa = 4.92GPTPVSALIHH340 pKa = 6.2AATMVASGVFLVARR354 pKa = 11.84TWPVFEE360 pKa = 5.01ASALARR366 pKa = 11.84TVMLAIGVVTALYY379 pKa = 10.2AAVVALAQTDD389 pKa = 3.69IKK391 pKa = 11.29KK392 pKa = 9.04MLAYY396 pKa = 9.55STISQLGFMFAALGAGAWGAAMFHH420 pKa = 6.46LTTHH424 pKa = 7.02AAFKK428 pKa = 10.94ALLFLAAGSVIHH440 pKa = 6.84GAGTQDD446 pKa = 3.19MRR448 pKa = 11.84EE449 pKa = 3.94MGGLAKK455 pKa = 10.66AMPVTAACWTAGALALSGWWPLSGFFSKK483 pKa = 10.79DD484 pKa = 3.1AIVAAVGHH492 pKa = 5.61VSSVASAALILTSALTALYY511 pKa = 9.79VWRR514 pKa = 11.84ATALAFLGSRR524 pKa = 11.84RR525 pKa = 11.84SDD527 pKa = 3.27QHH529 pKa = 7.29AHH531 pKa = 5.96EE532 pKa = 5.25SGIAMAAPLAVLAVLAVGLGWFGRR556 pKa = 11.84PFFEE560 pKa = 5.18LLSAEE565 pKa = 4.34EE566 pKa = 4.08AHH568 pKa = 6.68PAAATIALAVVAVAAGTAAAFAWWARR594 pKa = 11.84GRR596 pKa = 11.84EE597 pKa = 4.12APEE600 pKa = 4.05SGVCGSAWDD609 pKa = 3.66RR610 pKa = 11.84VRR612 pKa = 11.84AGFGVDD618 pKa = 3.05AALQAVTVRR627 pKa = 11.84PVLALARR634 pKa = 11.84WLDD637 pKa = 3.73DD638 pKa = 3.59VADD641 pKa = 3.76EE642 pKa = 5.39RR643 pKa = 11.84IIDD646 pKa = 3.81RR647 pKa = 11.84LAEE650 pKa = 4.34GSGKK654 pKa = 10.18LAVAAGGALAKK665 pKa = 10.12LQSGEE670 pKa = 3.9ADD672 pKa = 3.46VYY674 pKa = 11.45ASFVAVGFVVLAAIVLWAGGRR695 pKa = 3.53
MM1 pKa = 7.47RR2 pKa = 11.84AVLAGVVRR10 pKa = 11.84RR11 pKa = 11.84AQPARR16 pKa = 11.84HH17 pKa = 5.72HH18 pKa = 6.37RR19 pKa = 11.84GARR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84DD26 pKa = 3.18HH27 pKa = 6.62AGSRR31 pKa = 11.84KK32 pKa = 9.67RR33 pKa = 11.84PSCVAGRR40 pKa = 11.84CVRR43 pKa = 11.84RR44 pKa = 11.84VVWQRR49 pKa = 11.84AGDD52 pKa = 3.74RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.83GVAAGGSRR63 pKa = 11.84SCGGAVAPAFALPDD77 pKa = 3.43LRR79 pKa = 11.84QEE81 pKa = 3.81PRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84AEE87 pKa = 3.62RR88 pKa = 11.84GEE90 pKa = 4.04RR91 pKa = 11.84LMGALLVSIGLPLAITVVIVAGGARR116 pKa = 11.84SVRR119 pKa = 11.84VTRR122 pKa = 11.84VFALAGPLAALAAGASLIASAHH144 pKa = 5.92GGAAAPSWNGAWLAQGGAAAPVGFAGDD171 pKa = 3.46ALAALMLVIVGAVAACVVAFSWGYY195 pKa = 8.18MAHH198 pKa = 7.38EE199 pKa = 5.39DD200 pKa = 3.85GQPRR204 pKa = 11.84YY205 pKa = 9.73YY206 pKa = 10.75AVLMLFVSAMSVLVLADD223 pKa = 3.5NLVTLFIGWEE233 pKa = 3.97LVGACSYY240 pKa = 11.33LLIGFWYY247 pKa = 9.69RR248 pKa = 11.84KK249 pKa = 8.5PAATRR254 pKa = 11.84AAIKK258 pKa = 10.7AFMTTRR264 pKa = 11.84VGDD267 pKa = 3.54VAMLVGMAVLWRR279 pKa = 11.84HH280 pKa = 6.03AGTLSVRR287 pKa = 11.84EE288 pKa = 3.95IVAALPRR295 pKa = 11.84MEE297 pKa = 5.44PGVITAAALLLFGGAAGKK315 pKa = 9.66SAQFPLHH322 pKa = 5.89IWLPDD327 pKa = 3.24AMEE330 pKa = 4.92GPTPVSALIHH340 pKa = 6.2AATMVASGVFLVARR354 pKa = 11.84TWPVFEE360 pKa = 5.01ASALARR366 pKa = 11.84TVMLAIGVVTALYY379 pKa = 10.2AAVVALAQTDD389 pKa = 3.69IKK391 pKa = 11.29KK392 pKa = 9.04MLAYY396 pKa = 9.55STISQLGFMFAALGAGAWGAAMFHH420 pKa = 6.46LTTHH424 pKa = 7.02AAFKK428 pKa = 10.94ALLFLAAGSVIHH440 pKa = 6.84GAGTQDD446 pKa = 3.19MRR448 pKa = 11.84EE449 pKa = 3.94MGGLAKK455 pKa = 10.66AMPVTAACWTAGALALSGWWPLSGFFSKK483 pKa = 10.79DD484 pKa = 3.1AIVAAVGHH492 pKa = 5.61VSSVASAALILTSALTALYY511 pKa = 9.79VWRR514 pKa = 11.84ATALAFLGSRR524 pKa = 11.84RR525 pKa = 11.84SDD527 pKa = 3.27QHH529 pKa = 7.29AHH531 pKa = 5.96EE532 pKa = 5.25SGIAMAAPLAVLAVLAVGLGWFGRR556 pKa = 11.84PFFEE560 pKa = 5.18LLSAEE565 pKa = 4.34EE566 pKa = 4.08AHH568 pKa = 6.68PAAATIALAVVAVAAGTAAAFAWWARR594 pKa = 11.84GRR596 pKa = 11.84EE597 pKa = 4.12APEE600 pKa = 4.05SGVCGSAWDD609 pKa = 3.66RR610 pKa = 11.84VRR612 pKa = 11.84AGFGVDD618 pKa = 3.05AALQAVTVRR627 pKa = 11.84PVLALARR634 pKa = 11.84WLDD637 pKa = 3.73DD638 pKa = 3.59VADD641 pKa = 3.76EE642 pKa = 5.39RR643 pKa = 11.84IIDD646 pKa = 3.81RR647 pKa = 11.84LAEE650 pKa = 4.34GSGKK654 pKa = 10.18LAVAAGGALAKK665 pKa = 10.12LQSGEE670 pKa = 3.9ADD672 pKa = 3.46VYY674 pKa = 11.45ASFVAVGFVVLAAIVLWAGGRR695 pKa = 3.53
Molecular weight: 71.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
552506 |
34 |
2925 |
333.0 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.068 ± 0.088 | 1.077 ± 0.031 |
5.639 ± 0.037 | 6.77 ± 0.071 |
3.051 ± 0.033 | 8.553 ± 0.06 |
2.055 ± 0.035 | 4.349 ± 0.043 |
2.869 ± 0.054 | 9.679 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.162 ± 0.028 | 1.717 ± 0.028 |
5.074 ± 0.042 | 2.537 ± 0.035 |
8.008 ± 0.078 | 5.444 ± 0.045 |
5.069 ± 0.047 | 9.522 ± 0.057 |
1.105 ± 0.022 | 2.252 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
