
Pseudoxanthomonas sp. CF125
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Pseudoxanthomonas; unclassified Pseudoxanthomonas
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
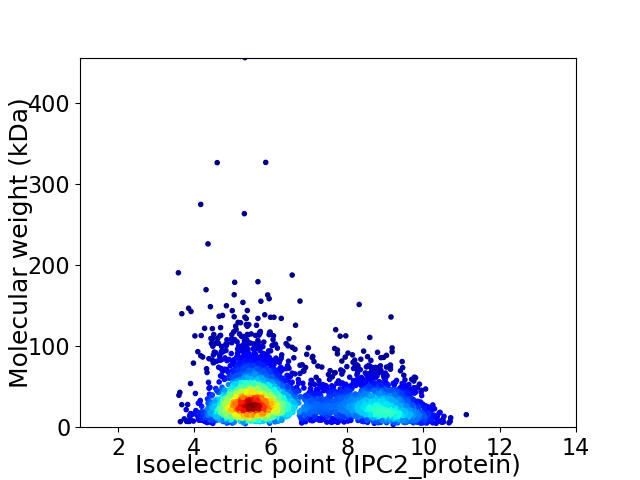
Virtual 2D-PAGE plot for 3691 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
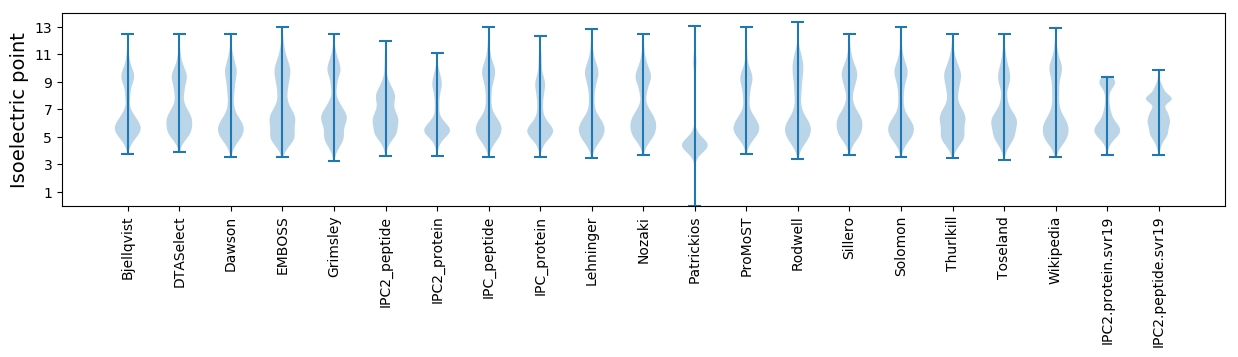
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1BTW0|A0A1H1BTW0_9GAMM ATP-binding protein Uup OS=Pseudoxanthomonas sp. CF125 OX=1855303 GN=uup PE=3 SV=1
MM1 pKa = 7.35NKK3 pKa = 9.03VFSLVWSRR11 pKa = 11.84SKK13 pKa = 10.61QALVVASEE21 pKa = 4.17LASAQGRR28 pKa = 11.84KK29 pKa = 8.61EE30 pKa = 3.69GRR32 pKa = 11.84PRR34 pKa = 11.84LRR36 pKa = 11.84DD37 pKa = 3.41ATLGAVLLGCGIAMALPAAHH57 pKa = 6.42AQEE60 pKa = 4.22LTIEE64 pKa = 4.29RR65 pKa = 11.84DD66 pKa = 3.17GDD68 pKa = 3.66VCTARR73 pKa = 11.84VEE75 pKa = 4.42GQAEE79 pKa = 4.34GRR81 pKa = 11.84QIDD84 pKa = 4.16CALFDD89 pKa = 3.74QANVGINAVVGSGYY103 pKa = 10.59FLADD107 pKa = 3.49GAGDD111 pKa = 3.95GSDD114 pKa = 4.25DD115 pKa = 3.8PTVSGDD121 pKa = 3.24GALAAGRR128 pKa = 11.84NSSALNNATTAVGAEE143 pKa = 4.16SAAVGLGASAFGQGAMAFGDD163 pKa = 3.56ASVAIGQSAYY173 pKa = 10.68ASGDD177 pKa = 3.67GPVMGLGNTAIGASASAEE195 pKa = 4.0AEE197 pKa = 4.02QGGTAVGALSLAAGTGSTAIGGNAQAYY224 pKa = 10.15GDD226 pKa = 3.97GSLVGGFGSGANGEE240 pKa = 4.29AGVALGANSRR250 pKa = 11.84ALGGDD255 pKa = 3.58FGVAIGGSSYY265 pKa = 11.22AIGQGATSVGGLAWAEE281 pKa = 4.18GNYY284 pKa = 10.56SLASGYY290 pKa = 11.02YY291 pKa = 7.79STAVGDD297 pKa = 3.67GSIAFGGGLGVDD309 pKa = 4.18LDD311 pKa = 4.31GDD313 pKa = 4.16GLIGPGEE320 pKa = 3.98ATIAAGYY327 pKa = 9.07HH328 pKa = 5.39AVALGYY334 pKa = 9.7QAAALGDD341 pKa = 3.75GTLALGDD348 pKa = 3.66QAQALQLNATAVGTMSVAAGEE369 pKa = 4.49SSTATGANSVATGAEE384 pKa = 4.07SSAYY388 pKa = 9.55GANSYY393 pKa = 11.58ADD395 pKa = 3.91ADD397 pKa = 3.86YY398 pKa = 9.88STAIGSGSLALGSGSTAIGAGANALGTEE426 pKa = 4.79SVASGYY432 pKa = 11.23GSVASGEE439 pKa = 3.97YY440 pKa = 10.4SIAQGSSSQALGEE453 pKa = 3.85ASIAIGVQSVASGTDD468 pKa = 3.31SIGIGAGSQAGDD480 pKa = 3.31FGTAVGTLSNATGYY494 pKa = 9.98GASAFGDD501 pKa = 3.85NAWATNDD508 pKa = 3.1FSLAMGANSTSLGIGGIAIGTDD530 pKa = 3.39SYY532 pKa = 11.89VGADD536 pKa = 3.58ASHH539 pKa = 7.02GIAIGNLVMANGKK552 pKa = 9.82NSIAMGGLWTTAGADD567 pKa = 3.44GAIAIGGTKK576 pKa = 10.14ACTSFMCIEE585 pKa = 4.37EE586 pKa = 4.2NTLAWGINSVALGAGATTQADD607 pKa = 3.99GATALGSGAQAGGIDD622 pKa = 4.32SISIGMDD629 pKa = 2.87SRR631 pKa = 11.84AGSVNSVGIGARR643 pKa = 11.84TYY645 pKa = 11.3ALGDD649 pKa = 3.34NSVALGADD657 pKa = 3.45SVTSRR662 pKa = 11.84ANTVTVGSEE671 pKa = 3.7WTGMTRR677 pKa = 11.84QITGVADD684 pKa = 3.46GTEE687 pKa = 4.2ANDD690 pKa = 4.3AVNLSQLNAVADD702 pKa = 4.22SVGDD706 pKa = 3.83ANDD709 pKa = 3.59YY710 pKa = 11.25LSVNGLGDD718 pKa = 3.59GTDD721 pKa = 3.11AAIAEE726 pKa = 4.57GEE728 pKa = 4.18NASAAGANAVAIGIEE743 pKa = 4.55SSALGYY749 pKa = 10.14QSNAVGDD756 pKa = 3.88YY757 pKa = 8.52STAIGGSSLALGAGSSALGTGASAIGDD784 pKa = 3.53YY785 pKa = 11.06SSANGYY791 pKa = 8.94GAIAEE796 pKa = 4.5GSNSNAFGANAAAIGNSSFAGGDD819 pKa = 3.43SAQANGGGSVALGASSIANGSNGVSVGAGAEE850 pKa = 4.36SNGFFAAAIGGYY862 pKa = 10.29ALADD866 pKa = 3.69SPFSVAIGGFATASGWQATAIGSGAFASGFNATATGAGAMAGDD909 pKa = 4.0GGVAIGAGAASGYY922 pKa = 8.87WFYY925 pKa = 11.65DD926 pKa = 3.02EE927 pKa = 4.43TTGEE931 pKa = 4.01NTFIGSSGTAVGQNAAATGMFSTALGGFSDD961 pKa = 5.13GSFASAGATGMYY973 pKa = 8.52STALGAGAYY982 pKa = 9.63AGSLDD987 pKa = 3.55GVVPGDD993 pKa = 3.42YY994 pKa = 7.56TTAVGAEE1001 pKa = 3.98SWAVGDD1007 pKa = 4.21SATSLGFNSYY1017 pKa = 10.09AQSANSTVVGANAWTTVDD1035 pKa = 3.73ADD1037 pKa = 3.52NSVALGEE1044 pKa = 4.44GSLADD1049 pKa = 3.81RR1050 pKa = 11.84ANTVSVGAAQDD1061 pKa = 3.22WTDD1064 pKa = 2.96AGGNVHH1070 pKa = 7.03AAIDD1074 pKa = 3.75RR1075 pKa = 11.84QITNVAAGTEE1085 pKa = 4.1ATDD1088 pKa = 4.2AVNLAQLNEE1097 pKa = 4.13VADD1100 pKa = 3.87NLGEE1104 pKa = 4.11FTQNAVAYY1112 pKa = 10.22DD1113 pKa = 4.18DD1114 pKa = 5.58DD1115 pKa = 4.56SHH1117 pKa = 7.72DD1118 pKa = 3.94KK1119 pKa = 10.13LTLEE1123 pKa = 4.52GAGGSTISNLAAGTQASDD1141 pKa = 3.25AVNVEE1146 pKa = 3.96QLNSVAAAFGGGASLGANGMLVAPTYY1172 pKa = 8.94TIQGGGYY1179 pKa = 10.1ANAGDD1184 pKa = 4.05AFAAVDD1190 pKa = 3.88GMLSGLNGRR1199 pKa = 11.84LTQIEE1204 pKa = 4.37QNLGNGGGIGGPPTGTGDD1222 pKa = 3.47GLAIGTGSNATDD1234 pKa = 3.53TTDD1237 pKa = 3.18TAVGNGANVGADD1249 pKa = 2.83NGTAVGNNSSIAASATDD1266 pKa = 3.74SVAIGADD1273 pKa = 3.25ASVTAASGTAIGQGATVTGAGSVALGQGSVADD1305 pKa = 3.71QANTVSVGSAGNEE1318 pKa = 3.28RR1319 pKa = 11.84RR1320 pKa = 11.84VTNVAAGVAATDD1332 pKa = 3.75AANVGQMQAGDD1343 pKa = 3.77AATLASANDD1352 pKa = 3.76YY1353 pKa = 10.61TDD1355 pKa = 3.19ATATEE1360 pKa = 4.33TLSAANAYY1368 pKa = 8.05TDD1370 pKa = 4.19SRR1372 pKa = 11.84LQTFDD1377 pKa = 4.96DD1378 pKa = 4.84DD1379 pKa = 4.27FTALRR1384 pKa = 11.84NDD1386 pKa = 2.91VDD1388 pKa = 4.22HH1389 pKa = 7.37RR1390 pKa = 11.84FSQQDD1395 pKa = 3.02RR1396 pKa = 11.84RR1397 pKa = 11.84IDD1399 pKa = 3.22KK1400 pKa = 9.68MGAMSSAMMNMSINAAGSRR1419 pKa = 11.84SPRR1422 pKa = 11.84GRR1424 pKa = 11.84IAVGAGWQNGEE1435 pKa = 4.1SALSVGYY1442 pKa = 9.95SKK1444 pKa = 11.0QIGTRR1449 pKa = 11.84ASFSIGGAFSSDD1461 pKa = 3.06EE1462 pKa = 3.96KK1463 pKa = 10.96SAGVGFGVDD1472 pKa = 3.19LL1473 pKa = 4.58
MM1 pKa = 7.35NKK3 pKa = 9.03VFSLVWSRR11 pKa = 11.84SKK13 pKa = 10.61QALVVASEE21 pKa = 4.17LASAQGRR28 pKa = 11.84KK29 pKa = 8.61EE30 pKa = 3.69GRR32 pKa = 11.84PRR34 pKa = 11.84LRR36 pKa = 11.84DD37 pKa = 3.41ATLGAVLLGCGIAMALPAAHH57 pKa = 6.42AQEE60 pKa = 4.22LTIEE64 pKa = 4.29RR65 pKa = 11.84DD66 pKa = 3.17GDD68 pKa = 3.66VCTARR73 pKa = 11.84VEE75 pKa = 4.42GQAEE79 pKa = 4.34GRR81 pKa = 11.84QIDD84 pKa = 4.16CALFDD89 pKa = 3.74QANVGINAVVGSGYY103 pKa = 10.59FLADD107 pKa = 3.49GAGDD111 pKa = 3.95GSDD114 pKa = 4.25DD115 pKa = 3.8PTVSGDD121 pKa = 3.24GALAAGRR128 pKa = 11.84NSSALNNATTAVGAEE143 pKa = 4.16SAAVGLGASAFGQGAMAFGDD163 pKa = 3.56ASVAIGQSAYY173 pKa = 10.68ASGDD177 pKa = 3.67GPVMGLGNTAIGASASAEE195 pKa = 4.0AEE197 pKa = 4.02QGGTAVGALSLAAGTGSTAIGGNAQAYY224 pKa = 10.15GDD226 pKa = 3.97GSLVGGFGSGANGEE240 pKa = 4.29AGVALGANSRR250 pKa = 11.84ALGGDD255 pKa = 3.58FGVAIGGSSYY265 pKa = 11.22AIGQGATSVGGLAWAEE281 pKa = 4.18GNYY284 pKa = 10.56SLASGYY290 pKa = 11.02YY291 pKa = 7.79STAVGDD297 pKa = 3.67GSIAFGGGLGVDD309 pKa = 4.18LDD311 pKa = 4.31GDD313 pKa = 4.16GLIGPGEE320 pKa = 3.98ATIAAGYY327 pKa = 9.07HH328 pKa = 5.39AVALGYY334 pKa = 9.7QAAALGDD341 pKa = 3.75GTLALGDD348 pKa = 3.66QAQALQLNATAVGTMSVAAGEE369 pKa = 4.49SSTATGANSVATGAEE384 pKa = 4.07SSAYY388 pKa = 9.55GANSYY393 pKa = 11.58ADD395 pKa = 3.91ADD397 pKa = 3.86YY398 pKa = 9.88STAIGSGSLALGSGSTAIGAGANALGTEE426 pKa = 4.79SVASGYY432 pKa = 11.23GSVASGEE439 pKa = 3.97YY440 pKa = 10.4SIAQGSSSQALGEE453 pKa = 3.85ASIAIGVQSVASGTDD468 pKa = 3.31SIGIGAGSQAGDD480 pKa = 3.31FGTAVGTLSNATGYY494 pKa = 9.98GASAFGDD501 pKa = 3.85NAWATNDD508 pKa = 3.1FSLAMGANSTSLGIGGIAIGTDD530 pKa = 3.39SYY532 pKa = 11.89VGADD536 pKa = 3.58ASHH539 pKa = 7.02GIAIGNLVMANGKK552 pKa = 9.82NSIAMGGLWTTAGADD567 pKa = 3.44GAIAIGGTKK576 pKa = 10.14ACTSFMCIEE585 pKa = 4.37EE586 pKa = 4.2NTLAWGINSVALGAGATTQADD607 pKa = 3.99GATALGSGAQAGGIDD622 pKa = 4.32SISIGMDD629 pKa = 2.87SRR631 pKa = 11.84AGSVNSVGIGARR643 pKa = 11.84TYY645 pKa = 11.3ALGDD649 pKa = 3.34NSVALGADD657 pKa = 3.45SVTSRR662 pKa = 11.84ANTVTVGSEE671 pKa = 3.7WTGMTRR677 pKa = 11.84QITGVADD684 pKa = 3.46GTEE687 pKa = 4.2ANDD690 pKa = 4.3AVNLSQLNAVADD702 pKa = 4.22SVGDD706 pKa = 3.83ANDD709 pKa = 3.59YY710 pKa = 11.25LSVNGLGDD718 pKa = 3.59GTDD721 pKa = 3.11AAIAEE726 pKa = 4.57GEE728 pKa = 4.18NASAAGANAVAIGIEE743 pKa = 4.55SSALGYY749 pKa = 10.14QSNAVGDD756 pKa = 3.88YY757 pKa = 8.52STAIGGSSLALGAGSSALGTGASAIGDD784 pKa = 3.53YY785 pKa = 11.06SSANGYY791 pKa = 8.94GAIAEE796 pKa = 4.5GSNSNAFGANAAAIGNSSFAGGDD819 pKa = 3.43SAQANGGGSVALGASSIANGSNGVSVGAGAEE850 pKa = 4.36SNGFFAAAIGGYY862 pKa = 10.29ALADD866 pKa = 3.69SPFSVAIGGFATASGWQATAIGSGAFASGFNATATGAGAMAGDD909 pKa = 4.0GGVAIGAGAASGYY922 pKa = 8.87WFYY925 pKa = 11.65DD926 pKa = 3.02EE927 pKa = 4.43TTGEE931 pKa = 4.01NTFIGSSGTAVGQNAAATGMFSTALGGFSDD961 pKa = 5.13GSFASAGATGMYY973 pKa = 8.52STALGAGAYY982 pKa = 9.63AGSLDD987 pKa = 3.55GVVPGDD993 pKa = 3.42YY994 pKa = 7.56TTAVGAEE1001 pKa = 3.98SWAVGDD1007 pKa = 4.21SATSLGFNSYY1017 pKa = 10.09AQSANSTVVGANAWTTVDD1035 pKa = 3.73ADD1037 pKa = 3.52NSVALGEE1044 pKa = 4.44GSLADD1049 pKa = 3.81RR1050 pKa = 11.84ANTVSVGAAQDD1061 pKa = 3.22WTDD1064 pKa = 2.96AGGNVHH1070 pKa = 7.03AAIDD1074 pKa = 3.75RR1075 pKa = 11.84QITNVAAGTEE1085 pKa = 4.1ATDD1088 pKa = 4.2AVNLAQLNEE1097 pKa = 4.13VADD1100 pKa = 3.87NLGEE1104 pKa = 4.11FTQNAVAYY1112 pKa = 10.22DD1113 pKa = 4.18DD1114 pKa = 5.58DD1115 pKa = 4.56SHH1117 pKa = 7.72DD1118 pKa = 3.94KK1119 pKa = 10.13LTLEE1123 pKa = 4.52GAGGSTISNLAAGTQASDD1141 pKa = 3.25AVNVEE1146 pKa = 3.96QLNSVAAAFGGGASLGANGMLVAPTYY1172 pKa = 8.94TIQGGGYY1179 pKa = 10.1ANAGDD1184 pKa = 4.05AFAAVDD1190 pKa = 3.88GMLSGLNGRR1199 pKa = 11.84LTQIEE1204 pKa = 4.37QNLGNGGGIGGPPTGTGDD1222 pKa = 3.47GLAIGTGSNATDD1234 pKa = 3.53TTDD1237 pKa = 3.18TAVGNGANVGADD1249 pKa = 2.83NGTAVGNNSSIAASATDD1266 pKa = 3.74SVAIGADD1273 pKa = 3.25ASVTAASGTAIGQGATVTGAGSVALGQGSVADD1305 pKa = 3.71QANTVSVGSAGNEE1318 pKa = 3.28RR1319 pKa = 11.84RR1320 pKa = 11.84VTNVAAGVAATDD1332 pKa = 3.75AANVGQMQAGDD1343 pKa = 3.77AATLASANDD1352 pKa = 3.76YY1353 pKa = 10.61TDD1355 pKa = 3.19ATATEE1360 pKa = 4.33TLSAANAYY1368 pKa = 8.05TDD1370 pKa = 4.19SRR1372 pKa = 11.84LQTFDD1377 pKa = 4.96DD1378 pKa = 4.84DD1379 pKa = 4.27FTALRR1384 pKa = 11.84NDD1386 pKa = 2.91VDD1388 pKa = 4.22HH1389 pKa = 7.37RR1390 pKa = 11.84FSQQDD1395 pKa = 3.02RR1396 pKa = 11.84RR1397 pKa = 11.84IDD1399 pKa = 3.22KK1400 pKa = 9.68MGAMSSAMMNMSINAAGSRR1419 pKa = 11.84SPRR1422 pKa = 11.84GRR1424 pKa = 11.84IAVGAGWQNGEE1435 pKa = 4.1SALSVGYY1442 pKa = 9.95SKK1444 pKa = 11.0QIGTRR1449 pKa = 11.84ASFSIGGAFSSDD1461 pKa = 3.06EE1462 pKa = 3.96KK1463 pKa = 10.96SAGVGFGVDD1472 pKa = 3.19LL1473 pKa = 4.58
Molecular weight: 139.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1GIX3|A0A1H1GIX3_9GAMM CubicO group peptidase beta-lactamase class C family OS=Pseudoxanthomonas sp. CF125 OX=1855303 GN=SAMN05216569_3235 PE=4 SV=1
MM1 pKa = 7.74SDD3 pKa = 3.9LVALTALEE11 pKa = 4.34PTTEE15 pKa = 4.37RR16 pKa = 11.84YY17 pKa = 8.9LAACWSDD24 pKa = 3.15STRR27 pKa = 11.84RR28 pKa = 11.84AYY30 pKa = 10.33AGDD33 pKa = 3.56LRR35 pKa = 11.84DD36 pKa = 3.59FLKK39 pKa = 10.52WGGCLPASEE48 pKa = 4.32QQVVEE53 pKa = 4.49YY54 pKa = 10.16INEE57 pKa = 3.89RR58 pKa = 11.84AKK60 pKa = 10.61VFRR63 pKa = 11.84VRR65 pKa = 11.84TLEE68 pKa = 3.77RR69 pKa = 11.84RR70 pKa = 11.84LTGIGMAHH78 pKa = 7.76ALQGHH83 pKa = 5.73TDD85 pKa = 3.62PTKK88 pKa = 9.72STLIKK93 pKa = 10.64KK94 pKa = 10.07LMRR97 pKa = 11.84GVKK100 pKa = 9.06RR101 pKa = 11.84VHH103 pKa = 6.13GVTPRR108 pKa = 11.84QAQPILHH115 pKa = 7.31ADD117 pKa = 4.14LQQMFEE123 pKa = 4.21RR124 pKa = 11.84TVGLTQTRR132 pKa = 11.84DD133 pKa = 3.07RR134 pKa = 11.84ALLMLGFSCAFRR146 pKa = 11.84RR147 pKa = 11.84SEE149 pKa = 4.18VVSLNVEE156 pKa = 4.03DD157 pKa = 5.82LSFTEE162 pKa = 4.4AGLTVRR168 pKa = 11.84LRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 10.88NDD175 pKa = 3.01QYY177 pKa = 11.92GKK179 pKa = 8.47TRR181 pKa = 11.84LIGVPHH187 pKa = 6.2GTGDD191 pKa = 3.4ACTVLAVRR199 pKa = 11.84AWLYY203 pKa = 11.24ASGIRR208 pKa = 11.84SGPLFRR214 pKa = 11.84RR215 pKa = 11.84LLKK218 pKa = 10.4SKK220 pKa = 10.63RR221 pKa = 11.84SGARR225 pKa = 11.84LSDD228 pKa = 3.33QSVSLIIKK236 pKa = 9.44RR237 pKa = 11.84YY238 pKa = 8.44AVAVGLPVDD247 pKa = 5.09RR248 pKa = 11.84ISGHH252 pKa = 5.18SLRR255 pKa = 11.84AGFVTSAVRR264 pKa = 11.84AGASLVSIQRR274 pKa = 11.84QTGHH278 pKa = 7.11ASLDD282 pKa = 3.19MLARR286 pKa = 11.84YY287 pKa = 9.07IRR289 pKa = 11.84EE290 pKa = 4.1LDD292 pKa = 3.61PFILNAHH299 pKa = 5.93DD300 pKa = 3.71HH301 pKa = 5.92LRR303 pKa = 4.37
MM1 pKa = 7.74SDD3 pKa = 3.9LVALTALEE11 pKa = 4.34PTTEE15 pKa = 4.37RR16 pKa = 11.84YY17 pKa = 8.9LAACWSDD24 pKa = 3.15STRR27 pKa = 11.84RR28 pKa = 11.84AYY30 pKa = 10.33AGDD33 pKa = 3.56LRR35 pKa = 11.84DD36 pKa = 3.59FLKK39 pKa = 10.52WGGCLPASEE48 pKa = 4.32QQVVEE53 pKa = 4.49YY54 pKa = 10.16INEE57 pKa = 3.89RR58 pKa = 11.84AKK60 pKa = 10.61VFRR63 pKa = 11.84VRR65 pKa = 11.84TLEE68 pKa = 3.77RR69 pKa = 11.84RR70 pKa = 11.84LTGIGMAHH78 pKa = 7.76ALQGHH83 pKa = 5.73TDD85 pKa = 3.62PTKK88 pKa = 9.72STLIKK93 pKa = 10.64KK94 pKa = 10.07LMRR97 pKa = 11.84GVKK100 pKa = 9.06RR101 pKa = 11.84VHH103 pKa = 6.13GVTPRR108 pKa = 11.84QAQPILHH115 pKa = 7.31ADD117 pKa = 4.14LQQMFEE123 pKa = 4.21RR124 pKa = 11.84TVGLTQTRR132 pKa = 11.84DD133 pKa = 3.07RR134 pKa = 11.84ALLMLGFSCAFRR146 pKa = 11.84RR147 pKa = 11.84SEE149 pKa = 4.18VVSLNVEE156 pKa = 4.03DD157 pKa = 5.82LSFTEE162 pKa = 4.4AGLTVRR168 pKa = 11.84LRR170 pKa = 11.84RR171 pKa = 11.84SKK173 pKa = 10.88NDD175 pKa = 3.01QYY177 pKa = 11.92GKK179 pKa = 8.47TRR181 pKa = 11.84LIGVPHH187 pKa = 6.2GTGDD191 pKa = 3.4ACTVLAVRR199 pKa = 11.84AWLYY203 pKa = 11.24ASGIRR208 pKa = 11.84SGPLFRR214 pKa = 11.84RR215 pKa = 11.84LLKK218 pKa = 10.4SKK220 pKa = 10.63RR221 pKa = 11.84SGARR225 pKa = 11.84LSDD228 pKa = 3.33QSVSLIIKK236 pKa = 9.44RR237 pKa = 11.84YY238 pKa = 8.44AVAVGLPVDD247 pKa = 5.09RR248 pKa = 11.84ISGHH252 pKa = 5.18SLRR255 pKa = 11.84AGFVTSAVRR264 pKa = 11.84AGASLVSIQRR274 pKa = 11.84QTGHH278 pKa = 7.11ASLDD282 pKa = 3.19MLARR286 pKa = 11.84YY287 pKa = 9.07IRR289 pKa = 11.84EE290 pKa = 4.1LDD292 pKa = 3.61PFILNAHH299 pKa = 5.93DD300 pKa = 3.71HH301 pKa = 5.92LRR303 pKa = 4.37
Molecular weight: 33.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1245779 |
31 |
4227 |
337.5 |
36.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.413 ± 0.057 | 0.805 ± 0.014 |
5.712 ± 0.026 | 5.474 ± 0.036 |
3.547 ± 0.025 | 8.599 ± 0.041 |
2.088 ± 0.019 | 4.369 ± 0.025 |
3.256 ± 0.035 | 10.498 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.02 | 2.818 ± 0.036 |
5.221 ± 0.03 | 3.744 ± 0.027 |
7.023 ± 0.045 | 5.728 ± 0.036 |
5.158 ± 0.053 | 7.248 ± 0.033 |
1.565 ± 0.02 | 2.543 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
