
Haliea sp. SAOS-164
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Haliea; unclassified Haliea
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
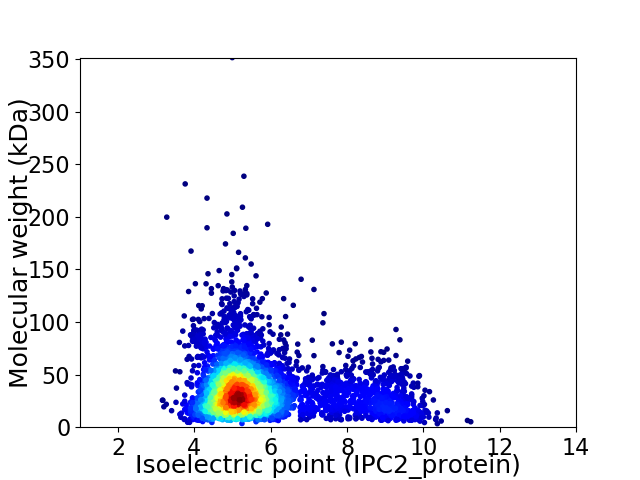
Virtual 2D-PAGE plot for 4187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0M9A0|A0A4Z0M9A0_9GAMM Glycerophosphodiester phosphodiesterase OS=Haliea sp. SAOS-164 OX=2562682 GN=glpQ PE=4 SV=1
MM1 pKa = 7.4SRR3 pKa = 11.84NLRR6 pKa = 11.84LLACTGVVLGLSSVYY21 pKa = 10.71SLPASGALSLSCSLPYY37 pKa = 9.91THH39 pKa = 7.26GKK41 pKa = 8.48PGKK44 pKa = 9.78DD45 pKa = 3.1CTGKK49 pKa = 8.43HH50 pKa = 5.31TIEE53 pKa = 3.91IQVAGGSPQTEE64 pKa = 4.2VQAEE68 pKa = 4.28LLALDD73 pKa = 4.19PNGVILGALPVEE85 pKa = 4.54NHH87 pKa = 6.82DD88 pKa = 4.01PAHH91 pKa = 5.81LRR93 pKa = 11.84SRR95 pKa = 11.84IDD97 pKa = 3.23EE98 pKa = 4.55DD99 pKa = 4.68DD100 pKa = 3.67YY101 pKa = 12.1SLRR104 pKa = 11.84TTNRR108 pKa = 11.84GVHH111 pKa = 5.29VVAAPIPLLSVTAVDD126 pKa = 3.98GDD128 pKa = 4.23EE129 pKa = 4.31TVHH132 pKa = 6.93AYY134 pKa = 10.62CSDD137 pKa = 3.32VPYY140 pKa = 10.97LRR142 pKa = 11.84LYY144 pKa = 10.5QPTGQVATAGDD155 pKa = 3.84TVDD158 pKa = 3.51VVLAATGLEE167 pKa = 4.14PASLQVFVDD176 pKa = 4.4GVEE179 pKa = 3.77IAAALGLTPATDD191 pKa = 4.12FPGGPYY197 pKa = 10.1AGNVSIGGEE206 pKa = 4.01NVEE209 pKa = 4.14ITDD212 pKa = 4.3LVVDD216 pKa = 5.02LGDD219 pKa = 3.6IEE221 pKa = 4.92EE222 pKa = 4.7PSSNTVSFTLEE233 pKa = 3.65GLPAGGHH240 pKa = 5.8VIFADD245 pKa = 4.07GEE247 pKa = 4.23RR248 pKa = 11.84LANRR252 pKa = 11.84PTRR255 pKa = 11.84GPVSDD260 pKa = 4.25ACHH263 pKa = 6.55IDD265 pKa = 3.46DD266 pKa = 5.43MEE268 pKa = 4.9DD269 pKa = 3.0AGQFAVFGVDD279 pKa = 2.72IEE281 pKa = 5.0SPFEE285 pKa = 3.97QQVVNAVPTPVSGVVKK301 pKa = 10.46HH302 pKa = 6.14GLPIAATKK310 pKa = 10.55INGAALDD317 pKa = 3.66VSGQSFVAGDD327 pKa = 3.82GKK329 pKa = 9.69FTADD333 pKa = 2.77QYY335 pKa = 11.53EE336 pKa = 4.06YY337 pKa = 10.66HH338 pKa = 6.9FDD340 pKa = 3.52EE341 pKa = 6.66AIPQTDD347 pKa = 3.41LRR349 pKa = 11.84ADD351 pKa = 4.16LDD353 pKa = 4.08AGSAPLGSFDD363 pKa = 4.43RR364 pKa = 11.84GSNRR368 pKa = 11.84LTVDD372 pKa = 3.57ALDD375 pKa = 3.92TYY377 pKa = 11.06GNRR380 pKa = 11.84AFDD383 pKa = 3.75EE384 pKa = 4.52LFFAVGGVQSPTAPLEE400 pKa = 4.11TLAANALGFNVEE412 pKa = 4.31RR413 pKa = 11.84SVSPLLMVALDD424 pKa = 3.84EE425 pKa = 4.92AEE427 pKa = 4.15EE428 pKa = 4.7AIQNSFVVGLTEE440 pKa = 6.07DD441 pKa = 5.07GIQALMDD448 pKa = 3.9QKK450 pKa = 11.3CPDD453 pKa = 3.36AGQAFKK459 pKa = 11.05EE460 pKa = 4.5SVQSKK465 pKa = 8.99IAEE468 pKa = 3.89IAPISKK474 pKa = 9.93KK475 pKa = 10.36ISGGCSCSPNVEE487 pKa = 3.7IGITGSSIDD496 pKa = 4.31ANAISCPVTFQDD508 pKa = 3.79GKK510 pKa = 10.12IAVSIDD516 pKa = 3.48LPDD519 pKa = 3.37VTVYY523 pKa = 10.61GSAYY527 pKa = 8.94GHH529 pKa = 6.69CKK531 pKa = 9.42DD532 pKa = 4.09TVAGVCVAEE541 pKa = 4.33TTVDD545 pKa = 3.1ITSTTTISDD554 pKa = 3.86LSLSFNITEE563 pKa = 4.35GQFLGDD569 pKa = 3.77PPEE572 pKa = 4.26EE573 pKa = 4.01PVFVTGTSSVSTSGGSDD590 pKa = 3.35VDD592 pKa = 4.93CLAEE596 pKa = 3.94VCNWAIEE603 pKa = 4.56GIVTIFTFGTVDD615 pKa = 4.59LDD617 pKa = 3.49LTPAIDD623 pKa = 3.57VSQVTEE629 pKa = 4.16FNQAIGASEE638 pKa = 4.75PDD640 pKa = 4.87PIDD643 pKa = 4.43LGDD646 pKa = 3.43IKK648 pKa = 10.33IDD650 pKa = 3.57EE651 pKa = 4.54EE652 pKa = 4.22EE653 pKa = 4.28VEE655 pKa = 4.42EE656 pKa = 4.87FGQASLGGALEE667 pKa = 4.34SVEE670 pKa = 3.85ITPQGFLATLSGNFEE685 pKa = 4.27TLAVDD690 pKa = 3.86TDD692 pKa = 3.94VQPTPGAVVDD702 pKa = 4.08VAPAPTMQIPNAGDD716 pKa = 3.44ATVLLSDD723 pKa = 4.01EE724 pKa = 5.08VINQLFSSMAEE735 pKa = 3.87SGGLKK740 pKa = 8.61TACQYY745 pKa = 10.05TDD747 pKa = 3.2KK748 pKa = 11.28SVDD751 pKa = 3.58DD752 pKa = 4.71VLPTDD757 pKa = 4.83CDD759 pKa = 3.94ALDD762 pKa = 3.89GAAASGMCHH771 pKa = 7.58AIRR774 pKa = 11.84GTDD777 pKa = 3.59CEE779 pKa = 4.42TLAGDD784 pKa = 5.36GITGTAIKK792 pKa = 10.46QGVCHH797 pKa = 6.33GASGDD802 pKa = 3.83TCSGIAGSDD811 pKa = 3.67LEE813 pKa = 5.25RR814 pKa = 11.84NTCSGTPPTTLAAYY828 pKa = 9.29NQVLFCSRR836 pKa = 11.84QDD838 pKa = 3.34NPPRR842 pKa = 11.84FLIQDD847 pKa = 3.78DD848 pKa = 4.29AGTPATVEE856 pKa = 3.98TAVRR860 pKa = 11.84LSDD863 pKa = 3.61LSVAIVVDD871 pKa = 3.92RR872 pKa = 11.84DD873 pKa = 3.23NDD875 pKa = 3.69GFAEE879 pKa = 4.55LNDD882 pKa = 3.89TPDD885 pKa = 3.88CFGSDD890 pKa = 2.98ASVSGDD896 pKa = 3.16CALYY900 pKa = 8.39NTCLDD905 pKa = 4.14LNLVTTSSIATEE917 pKa = 4.25NLDD920 pKa = 3.89CPAGVPGFVTDD931 pKa = 3.9VVDD934 pKa = 4.3FQILNRR940 pKa = 11.84QEE942 pKa = 4.6GVVCGAAQEE951 pKa = 4.55TVDD954 pKa = 3.82GQLVGTSNDD963 pKa = 3.8DD964 pKa = 3.54DD965 pKa = 4.1TVDD968 pKa = 5.3LIGEE972 pKa = 4.33NVDD975 pKa = 3.51TYY977 pKa = 11.68APPLCVEE984 pKa = 4.3GLDD987 pKa = 3.59LGGFVTLQNPKK998 pKa = 9.9IIAVEE1003 pKa = 4.02TDD1005 pKa = 2.67GDD1007 pKa = 4.11ADD1009 pKa = 3.64FQEE1012 pKa = 4.97YY1013 pKa = 10.26IGITGEE1019 pKa = 4.03VAAPP1023 pKa = 3.49
MM1 pKa = 7.4SRR3 pKa = 11.84NLRR6 pKa = 11.84LLACTGVVLGLSSVYY21 pKa = 10.71SLPASGALSLSCSLPYY37 pKa = 9.91THH39 pKa = 7.26GKK41 pKa = 8.48PGKK44 pKa = 9.78DD45 pKa = 3.1CTGKK49 pKa = 8.43HH50 pKa = 5.31TIEE53 pKa = 3.91IQVAGGSPQTEE64 pKa = 4.2VQAEE68 pKa = 4.28LLALDD73 pKa = 4.19PNGVILGALPVEE85 pKa = 4.54NHH87 pKa = 6.82DD88 pKa = 4.01PAHH91 pKa = 5.81LRR93 pKa = 11.84SRR95 pKa = 11.84IDD97 pKa = 3.23EE98 pKa = 4.55DD99 pKa = 4.68DD100 pKa = 3.67YY101 pKa = 12.1SLRR104 pKa = 11.84TTNRR108 pKa = 11.84GVHH111 pKa = 5.29VVAAPIPLLSVTAVDD126 pKa = 3.98GDD128 pKa = 4.23EE129 pKa = 4.31TVHH132 pKa = 6.93AYY134 pKa = 10.62CSDD137 pKa = 3.32VPYY140 pKa = 10.97LRR142 pKa = 11.84LYY144 pKa = 10.5QPTGQVATAGDD155 pKa = 3.84TVDD158 pKa = 3.51VVLAATGLEE167 pKa = 4.14PASLQVFVDD176 pKa = 4.4GVEE179 pKa = 3.77IAAALGLTPATDD191 pKa = 4.12FPGGPYY197 pKa = 10.1AGNVSIGGEE206 pKa = 4.01NVEE209 pKa = 4.14ITDD212 pKa = 4.3LVVDD216 pKa = 5.02LGDD219 pKa = 3.6IEE221 pKa = 4.92EE222 pKa = 4.7PSSNTVSFTLEE233 pKa = 3.65GLPAGGHH240 pKa = 5.8VIFADD245 pKa = 4.07GEE247 pKa = 4.23RR248 pKa = 11.84LANRR252 pKa = 11.84PTRR255 pKa = 11.84GPVSDD260 pKa = 4.25ACHH263 pKa = 6.55IDD265 pKa = 3.46DD266 pKa = 5.43MEE268 pKa = 4.9DD269 pKa = 3.0AGQFAVFGVDD279 pKa = 2.72IEE281 pKa = 5.0SPFEE285 pKa = 3.97QQVVNAVPTPVSGVVKK301 pKa = 10.46HH302 pKa = 6.14GLPIAATKK310 pKa = 10.55INGAALDD317 pKa = 3.66VSGQSFVAGDD327 pKa = 3.82GKK329 pKa = 9.69FTADD333 pKa = 2.77QYY335 pKa = 11.53EE336 pKa = 4.06YY337 pKa = 10.66HH338 pKa = 6.9FDD340 pKa = 3.52EE341 pKa = 6.66AIPQTDD347 pKa = 3.41LRR349 pKa = 11.84ADD351 pKa = 4.16LDD353 pKa = 4.08AGSAPLGSFDD363 pKa = 4.43RR364 pKa = 11.84GSNRR368 pKa = 11.84LTVDD372 pKa = 3.57ALDD375 pKa = 3.92TYY377 pKa = 11.06GNRR380 pKa = 11.84AFDD383 pKa = 3.75EE384 pKa = 4.52LFFAVGGVQSPTAPLEE400 pKa = 4.11TLAANALGFNVEE412 pKa = 4.31RR413 pKa = 11.84SVSPLLMVALDD424 pKa = 3.84EE425 pKa = 4.92AEE427 pKa = 4.15EE428 pKa = 4.7AIQNSFVVGLTEE440 pKa = 6.07DD441 pKa = 5.07GIQALMDD448 pKa = 3.9QKK450 pKa = 11.3CPDD453 pKa = 3.36AGQAFKK459 pKa = 11.05EE460 pKa = 4.5SVQSKK465 pKa = 8.99IAEE468 pKa = 3.89IAPISKK474 pKa = 9.93KK475 pKa = 10.36ISGGCSCSPNVEE487 pKa = 3.7IGITGSSIDD496 pKa = 4.31ANAISCPVTFQDD508 pKa = 3.79GKK510 pKa = 10.12IAVSIDD516 pKa = 3.48LPDD519 pKa = 3.37VTVYY523 pKa = 10.61GSAYY527 pKa = 8.94GHH529 pKa = 6.69CKK531 pKa = 9.42DD532 pKa = 4.09TVAGVCVAEE541 pKa = 4.33TTVDD545 pKa = 3.1ITSTTTISDD554 pKa = 3.86LSLSFNITEE563 pKa = 4.35GQFLGDD569 pKa = 3.77PPEE572 pKa = 4.26EE573 pKa = 4.01PVFVTGTSSVSTSGGSDD590 pKa = 3.35VDD592 pKa = 4.93CLAEE596 pKa = 3.94VCNWAIEE603 pKa = 4.56GIVTIFTFGTVDD615 pKa = 4.59LDD617 pKa = 3.49LTPAIDD623 pKa = 3.57VSQVTEE629 pKa = 4.16FNQAIGASEE638 pKa = 4.75PDD640 pKa = 4.87PIDD643 pKa = 4.43LGDD646 pKa = 3.43IKK648 pKa = 10.33IDD650 pKa = 3.57EE651 pKa = 4.54EE652 pKa = 4.22EE653 pKa = 4.28VEE655 pKa = 4.42EE656 pKa = 4.87FGQASLGGALEE667 pKa = 4.34SVEE670 pKa = 3.85ITPQGFLATLSGNFEE685 pKa = 4.27TLAVDD690 pKa = 3.86TDD692 pKa = 3.94VQPTPGAVVDD702 pKa = 4.08VAPAPTMQIPNAGDD716 pKa = 3.44ATVLLSDD723 pKa = 4.01EE724 pKa = 5.08VINQLFSSMAEE735 pKa = 3.87SGGLKK740 pKa = 8.61TACQYY745 pKa = 10.05TDD747 pKa = 3.2KK748 pKa = 11.28SVDD751 pKa = 3.58DD752 pKa = 4.71VLPTDD757 pKa = 4.83CDD759 pKa = 3.94ALDD762 pKa = 3.89GAAASGMCHH771 pKa = 7.58AIRR774 pKa = 11.84GTDD777 pKa = 3.59CEE779 pKa = 4.42TLAGDD784 pKa = 5.36GITGTAIKK792 pKa = 10.46QGVCHH797 pKa = 6.33GASGDD802 pKa = 3.83TCSGIAGSDD811 pKa = 3.67LEE813 pKa = 5.25RR814 pKa = 11.84NTCSGTPPTTLAAYY828 pKa = 9.29NQVLFCSRR836 pKa = 11.84QDD838 pKa = 3.34NPPRR842 pKa = 11.84FLIQDD847 pKa = 3.78DD848 pKa = 4.29AGTPATVEE856 pKa = 3.98TAVRR860 pKa = 11.84LSDD863 pKa = 3.61LSVAIVVDD871 pKa = 3.92RR872 pKa = 11.84DD873 pKa = 3.23NDD875 pKa = 3.69GFAEE879 pKa = 4.55LNDD882 pKa = 3.89TPDD885 pKa = 3.88CFGSDD890 pKa = 2.98ASVSGDD896 pKa = 3.16CALYY900 pKa = 8.39NTCLDD905 pKa = 4.14LNLVTTSSIATEE917 pKa = 4.25NLDD920 pKa = 3.89CPAGVPGFVTDD931 pKa = 3.9VVDD934 pKa = 4.3FQILNRR940 pKa = 11.84QEE942 pKa = 4.6GVVCGAAQEE951 pKa = 4.55TVDD954 pKa = 3.82GQLVGTSNDD963 pKa = 3.8DD964 pKa = 3.54DD965 pKa = 4.1TVDD968 pKa = 5.3LIGEE972 pKa = 4.33NVDD975 pKa = 3.51TYY977 pKa = 11.68APPLCVEE984 pKa = 4.3GLDD987 pKa = 3.59LGGFVTLQNPKK998 pKa = 9.9IIAVEE1003 pKa = 4.02TDD1005 pKa = 2.67GDD1007 pKa = 4.11ADD1009 pKa = 3.64FQEE1012 pKa = 4.97YY1013 pKa = 10.26IGITGEE1019 pKa = 4.03VAAPP1023 pKa = 3.49
Molecular weight: 105.8 kDa
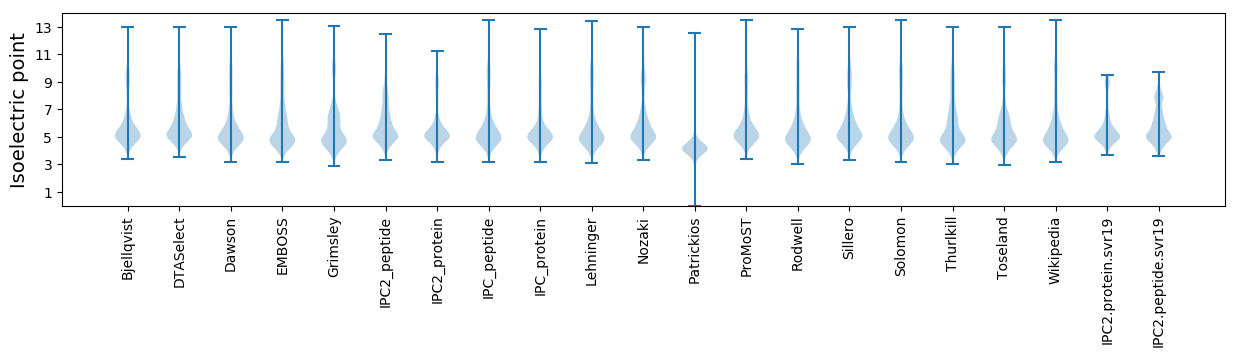
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0M0I0|A0A4Z0M0I0_9GAMM Methyltransferase OS=Haliea sp. SAOS-164 OX=2562682 GN=E4634_12215 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444718 |
26 |
3160 |
345.0 |
37.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.639 ± 0.056 | 1.115 ± 0.013 |
6.034 ± 0.028 | 6.36 ± 0.034 |
3.612 ± 0.022 | 8.242 ± 0.035 |
2.185 ± 0.018 | 4.529 ± 0.026 |
2.616 ± 0.029 | 10.87 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.018 | 2.8 ± 0.02 |
5.032 ± 0.026 | 3.887 ± 0.02 |
6.925 ± 0.037 | 5.607 ± 0.029 |
4.807 ± 0.027 | 7.216 ± 0.028 |
1.433 ± 0.014 | 2.789 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
