
Medicago truncatula (Barrel medic) (Medicago tribuloides)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade; NPAAA clade; Hologalegina;
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
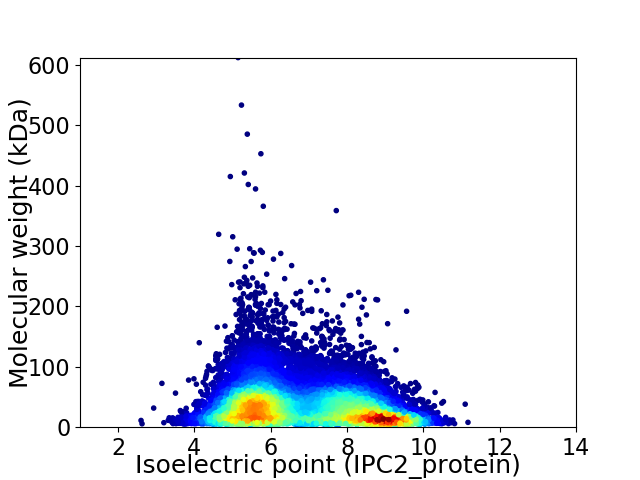
Virtual 2D-PAGE plot for 57067 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
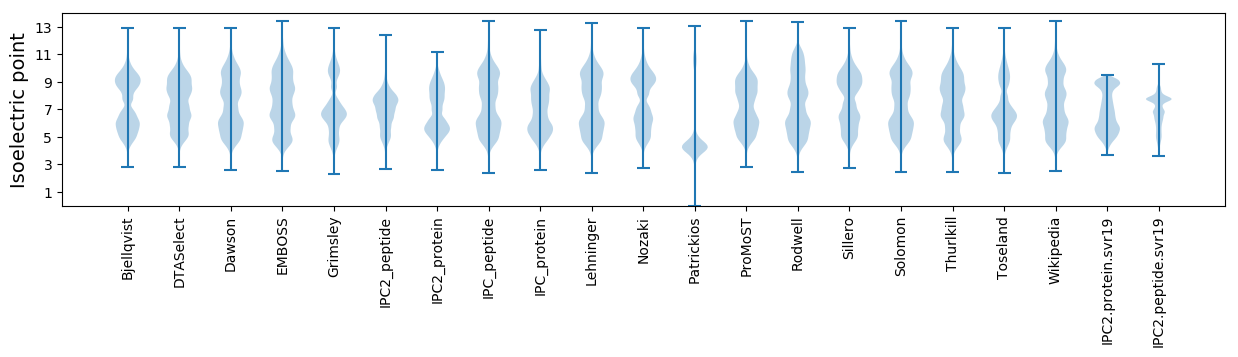
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G7LHX6|G7LHX6_MEDTR Putative transcription factor C2H2 family OS=Medicago truncatula OX=3880 GN=MTR_8g085530 PE=4 SV=2
MM1 pKa = 7.94DD2 pKa = 6.14PYY4 pKa = 11.39FNLYY8 pKa = 10.66DD9 pKa = 3.58IFQKK13 pKa = 10.71VSTRR17 pKa = 11.84ILDD20 pKa = 3.64TVLYY24 pKa = 9.71CIRR27 pKa = 11.84DD28 pKa = 3.7CAKK31 pKa = 10.69EE32 pKa = 3.88MVALNVEE39 pKa = 4.37GRR41 pKa = 11.84DD42 pKa = 3.49ILDD45 pKa = 3.51MKK47 pKa = 11.06VSLHH51 pKa = 5.07VNPYY55 pKa = 9.73IVEE58 pKa = 4.64DD59 pKa = 4.02YY60 pKa = 10.76PFEE63 pKa = 3.97QSHH66 pKa = 6.78RR67 pKa = 11.84NLHH70 pKa = 5.68DD71 pKa = 3.4QQNEE75 pKa = 3.88DD76 pKa = 4.16SLDD79 pKa = 3.72EE80 pKa = 4.76LEE82 pKa = 5.89GDD84 pKa = 4.12DD85 pKa = 5.3LLHH88 pKa = 6.63EE89 pKa = 4.56LEE91 pKa = 4.87EE92 pKa = 5.87DD93 pKa = 3.88DD94 pKa = 6.59FLLEE98 pKa = 3.95LQEE101 pKa = 5.28DD102 pKa = 3.99HH103 pKa = 7.54FEE105 pKa = 4.46QNPHH109 pKa = 6.37NYY111 pKa = 9.71NDD113 pKa = 3.68QQSLDD118 pKa = 3.67FSDD121 pKa = 3.25QLEE124 pKa = 4.47YY125 pKa = 11.33NDD127 pKa = 5.74DD128 pKa = 3.86LLEE131 pKa = 4.28EE132 pKa = 4.7LGEE135 pKa = 4.01EE136 pKa = 4.79DD137 pKa = 5.55RR138 pKa = 11.84FEE140 pKa = 4.28QYY142 pKa = 10.56LRR144 pKa = 11.84NYY146 pKa = 10.45NDD148 pKa = 3.2QQTVDD153 pKa = 4.12LLEE156 pKa = 4.34EE157 pKa = 4.69LEE159 pKa = 5.37DD160 pKa = 5.28DD161 pKa = 5.28DD162 pKa = 6.0FLHH165 pKa = 6.16EE166 pKa = 4.51QYY168 pKa = 10.97PHH170 pKa = 7.46IYY172 pKa = 10.33NDD174 pKa = 3.49QQNVDD179 pKa = 4.9LLDD182 pKa = 3.49QLEE185 pKa = 4.7DD186 pKa = 3.82DD187 pKa = 4.89DD188 pKa = 6.57FVEE191 pKa = 4.62EE192 pKa = 4.31WEE194 pKa = 4.62EE195 pKa = 4.11DD196 pKa = 3.72DD197 pKa = 5.09SLHH200 pKa = 6.28EE201 pKa = 4.17LEE203 pKa = 4.99EE204 pKa = 4.21DD205 pKa = 3.6HH206 pKa = 7.54FEE208 pKa = 4.87HH209 pKa = 7.44NYY211 pKa = 10.57NDD213 pKa = 3.71QQNVDD218 pKa = 4.9LLDD221 pKa = 3.54QLEE224 pKa = 4.68DD225 pKa = 4.15DD226 pKa = 4.67NLLQEE231 pKa = 4.89LEE233 pKa = 4.16EE234 pKa = 4.37DD235 pKa = 3.73HH236 pKa = 7.15FGQYY240 pKa = 7.87PHH242 pKa = 7.38NYY244 pKa = 8.58IDD246 pKa = 3.71QQNMDD251 pKa = 5.01LLDD254 pKa = 3.59QLEE257 pKa = 4.61DD258 pKa = 4.15DD259 pKa = 5.18DD260 pKa = 5.83LLEE263 pKa = 4.42EE264 pKa = 4.89LEE266 pKa = 4.95EE267 pKa = 6.15DD268 pKa = 5.48DD269 pKa = 6.37SLQNWKK275 pKa = 10.43KK276 pKa = 10.27IILSIIIMINKK287 pKa = 9.1IWICNINNSSFQSLLKK303 pKa = 10.54LYY305 pKa = 10.38VSRR308 pKa = 11.84IMM310 pKa = 4.86
MM1 pKa = 7.94DD2 pKa = 6.14PYY4 pKa = 11.39FNLYY8 pKa = 10.66DD9 pKa = 3.58IFQKK13 pKa = 10.71VSTRR17 pKa = 11.84ILDD20 pKa = 3.64TVLYY24 pKa = 9.71CIRR27 pKa = 11.84DD28 pKa = 3.7CAKK31 pKa = 10.69EE32 pKa = 3.88MVALNVEE39 pKa = 4.37GRR41 pKa = 11.84DD42 pKa = 3.49ILDD45 pKa = 3.51MKK47 pKa = 11.06VSLHH51 pKa = 5.07VNPYY55 pKa = 9.73IVEE58 pKa = 4.64DD59 pKa = 4.02YY60 pKa = 10.76PFEE63 pKa = 3.97QSHH66 pKa = 6.78RR67 pKa = 11.84NLHH70 pKa = 5.68DD71 pKa = 3.4QQNEE75 pKa = 3.88DD76 pKa = 4.16SLDD79 pKa = 3.72EE80 pKa = 4.76LEE82 pKa = 5.89GDD84 pKa = 4.12DD85 pKa = 5.3LLHH88 pKa = 6.63EE89 pKa = 4.56LEE91 pKa = 4.87EE92 pKa = 5.87DD93 pKa = 3.88DD94 pKa = 6.59FLLEE98 pKa = 3.95LQEE101 pKa = 5.28DD102 pKa = 3.99HH103 pKa = 7.54FEE105 pKa = 4.46QNPHH109 pKa = 6.37NYY111 pKa = 9.71NDD113 pKa = 3.68QQSLDD118 pKa = 3.67FSDD121 pKa = 3.25QLEE124 pKa = 4.47YY125 pKa = 11.33NDD127 pKa = 5.74DD128 pKa = 3.86LLEE131 pKa = 4.28EE132 pKa = 4.7LGEE135 pKa = 4.01EE136 pKa = 4.79DD137 pKa = 5.55RR138 pKa = 11.84FEE140 pKa = 4.28QYY142 pKa = 10.56LRR144 pKa = 11.84NYY146 pKa = 10.45NDD148 pKa = 3.2QQTVDD153 pKa = 4.12LLEE156 pKa = 4.34EE157 pKa = 4.69LEE159 pKa = 5.37DD160 pKa = 5.28DD161 pKa = 5.28DD162 pKa = 6.0FLHH165 pKa = 6.16EE166 pKa = 4.51QYY168 pKa = 10.97PHH170 pKa = 7.46IYY172 pKa = 10.33NDD174 pKa = 3.49QQNVDD179 pKa = 4.9LLDD182 pKa = 3.49QLEE185 pKa = 4.7DD186 pKa = 3.82DD187 pKa = 4.89DD188 pKa = 6.57FVEE191 pKa = 4.62EE192 pKa = 4.31WEE194 pKa = 4.62EE195 pKa = 4.11DD196 pKa = 3.72DD197 pKa = 5.09SLHH200 pKa = 6.28EE201 pKa = 4.17LEE203 pKa = 4.99EE204 pKa = 4.21DD205 pKa = 3.6HH206 pKa = 7.54FEE208 pKa = 4.87HH209 pKa = 7.44NYY211 pKa = 10.57NDD213 pKa = 3.71QQNVDD218 pKa = 4.9LLDD221 pKa = 3.54QLEE224 pKa = 4.68DD225 pKa = 4.15DD226 pKa = 4.67NLLQEE231 pKa = 4.89LEE233 pKa = 4.16EE234 pKa = 4.37DD235 pKa = 3.73HH236 pKa = 7.15FGQYY240 pKa = 7.87PHH242 pKa = 7.38NYY244 pKa = 8.58IDD246 pKa = 3.71QQNMDD251 pKa = 5.01LLDD254 pKa = 3.59QLEE257 pKa = 4.61DD258 pKa = 4.15DD259 pKa = 5.18DD260 pKa = 5.83LLEE263 pKa = 4.42EE264 pKa = 4.89LEE266 pKa = 4.95EE267 pKa = 6.15DD268 pKa = 5.48DD269 pKa = 6.37SLQNWKK275 pKa = 10.43KK276 pKa = 10.27IILSIIIMINKK287 pKa = 9.1IWICNINNSSFQSLLKK303 pKa = 10.54LYY305 pKa = 10.38VSRR308 pKa = 11.84IMM310 pKa = 4.86
Molecular weight: 37.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G7J4N2|G7J4N2_MEDTR Uncharacterized protein OS=Medicago truncatula OX=3880 GN=MTR_3g113040 PE=4 SV=1
MM1 pKa = 7.9SIPNIGSKK9 pKa = 10.01KK10 pKa = 9.92KK11 pKa = 10.39KK12 pKa = 10.25NNNQVVWAPVARR24 pKa = 11.84SPFQGRR30 pKa = 11.84IRR32 pKa = 11.84GIPGSISRR40 pKa = 11.84GNNTWPVHH48 pKa = 4.93MPLRR52 pKa = 11.84MSRR55 pKa = 11.84ISRR58 pKa = 11.84SPLMGRR64 pKa = 11.84KK65 pKa = 8.6PVRR68 pKa = 11.84KK69 pKa = 9.89LKK71 pKa = 10.8
MM1 pKa = 7.9SIPNIGSKK9 pKa = 10.01KK10 pKa = 9.92KK11 pKa = 10.39KK12 pKa = 10.25NNNQVVWAPVARR24 pKa = 11.84SPFQGRR30 pKa = 11.84IRR32 pKa = 11.84GIPGSISRR40 pKa = 11.84GNNTWPVHH48 pKa = 4.93MPLRR52 pKa = 11.84MSRR55 pKa = 11.84ISRR58 pKa = 11.84SPLMGRR64 pKa = 11.84KK65 pKa = 8.6PVRR68 pKa = 11.84KK69 pKa = 9.89LKK71 pKa = 10.8
Molecular weight: 8.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
19735905 |
16 |
5385 |
345.8 |
38.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.81 ± 0.009 | 1.951 ± 0.005 |
5.298 ± 0.007 | 6.305 ± 0.012 |
4.508 ± 0.008 | 6.105 ± 0.01 |
2.498 ± 0.005 | 5.893 ± 0.009 |
6.526 ± 0.01 | 9.841 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.005 | 5.098 ± 0.008 |
4.607 ± 0.011 | 3.623 ± 0.008 |
4.84 ± 0.008 | 8.857 ± 0.011 |
5.062 ± 0.007 | 6.498 ± 0.007 |
1.282 ± 0.004 | 2.934 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
