
Sporomusa acidovorans DSM 3132
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Sporomusa; Sporomusa acidovorans
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
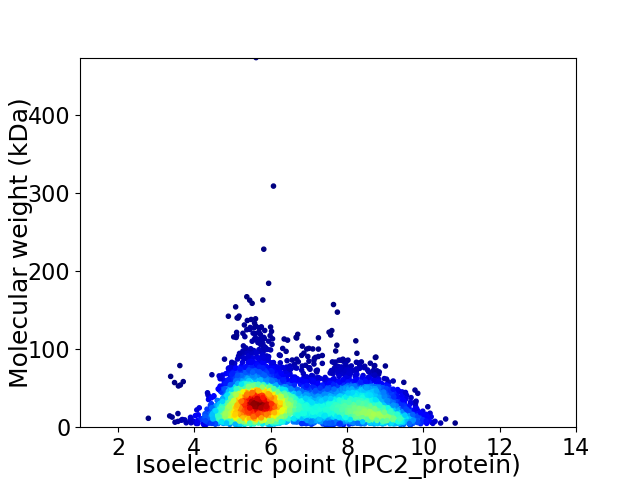
Virtual 2D-PAGE plot for 5767 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A259UWA1|A0A259UWA1_9FIRM Uncharacterized protein OS=Sporomusa acidovorans DSM 3132 OX=1123286 GN=SPACI_14860 PE=4 SV=1
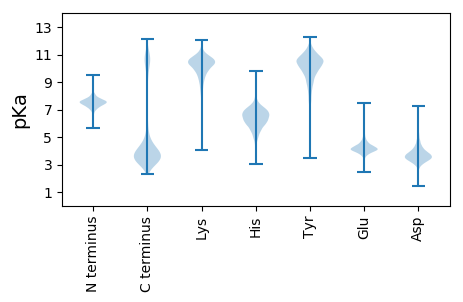
MM1 pKa = 7.74ASTFAGLSIANRR13 pKa = 11.84GLTAAQVGLTVTTNNMSNIDD33 pKa = 3.34TDD35 pKa = 4.26GYY37 pKa = 10.12SRR39 pKa = 11.84QVVSQTSIGPAAVYY53 pKa = 10.45SSSLVGNGVEE63 pKa = 4.26VTSVDD68 pKa = 3.23RR69 pKa = 11.84VNSFRR74 pKa = 11.84LNQKK78 pKa = 8.46YY79 pKa = 7.33WQEE82 pKa = 3.82NSSASEE88 pKa = 4.24LEE90 pKa = 4.14VTSNYY95 pKa = 8.47MEE97 pKa = 5.26KK98 pKa = 9.87IEE100 pKa = 5.1DD101 pKa = 3.66VFGTTDD107 pKa = 2.89SSDD110 pKa = 2.92ISEE113 pKa = 4.56ALDD116 pKa = 3.34TFEE119 pKa = 6.81AEE121 pKa = 4.99LEE123 pKa = 4.27TLADD127 pKa = 4.08DD128 pKa = 4.24PTNEE132 pKa = 4.42SIRR135 pKa = 11.84KK136 pKa = 8.83IVLSYY141 pKa = 11.2AKK143 pKa = 10.18SFCDD147 pKa = 3.6TLNTAAEE154 pKa = 4.29DD155 pKa = 3.85LEE157 pKa = 4.82SIRR160 pKa = 11.84DD161 pKa = 3.71QLNTEE166 pKa = 3.92VNTAVEE172 pKa = 4.88EE173 pKa = 4.31INSYY177 pKa = 10.87ASQIAEE183 pKa = 4.49LNRR186 pKa = 11.84QISTASASGASTNEE200 pKa = 4.04LEE202 pKa = 4.45DD203 pKa = 3.51QRR205 pKa = 11.84DD206 pKa = 3.92VIVDD210 pKa = 3.65KK211 pKa = 11.37LSGLVGISVTEE222 pKa = 4.4ADD224 pKa = 4.09DD225 pKa = 3.47STYY228 pKa = 10.57TITVDD233 pKa = 3.73GVTLVKK239 pKa = 10.57GDD241 pKa = 3.73EE242 pKa = 4.17ANEE245 pKa = 4.11LEE247 pKa = 5.3CYY249 pKa = 9.12TVTDD253 pKa = 4.24SDD255 pKa = 3.87SDD257 pKa = 4.01SYY259 pKa = 11.94GMYY262 pKa = 10.22GIRR265 pKa = 11.84WAEE268 pKa = 3.87SGKK271 pKa = 10.48DD272 pKa = 3.59FDD274 pKa = 5.14SGDD277 pKa = 3.75SGSLTGYY284 pKa = 10.23LQMRR288 pKa = 11.84DD289 pKa = 3.22GSTADD294 pKa = 3.25SKK296 pKa = 11.65GVVYY300 pKa = 10.13YY301 pKa = 9.66MNQLDD306 pKa = 4.5KK307 pKa = 10.73FAQTFAQAFNEE318 pKa = 4.54GVTVTSDD325 pKa = 2.66SGTEE329 pKa = 3.9TTYY332 pKa = 10.85SGHH335 pKa = 5.85VDD337 pKa = 3.35GVGIDD342 pKa = 3.95DD343 pKa = 5.96DD344 pKa = 4.23EE345 pKa = 4.63TTGIRR350 pKa = 11.84FFTYY354 pKa = 10.77DD355 pKa = 4.0DD356 pKa = 3.6VSSADD361 pKa = 3.83FVDD364 pKa = 3.8SGSDD368 pKa = 3.57LEE370 pKa = 4.25EE371 pKa = 4.77AYY373 pKa = 11.41SNITAANISVSKK385 pKa = 10.42DD386 pKa = 3.14IQDD389 pKa = 3.84DD390 pKa = 3.96TTKK393 pKa = 10.42IAASSTAGEE402 pKa = 4.22EE403 pKa = 4.57SNTDD407 pKa = 3.48VLDD410 pKa = 4.31DD411 pKa = 5.74LIDD414 pKa = 3.58ICSDD418 pKa = 3.44VNISGNSTATEE429 pKa = 4.17MYY431 pKa = 9.55NLIVATVAGDD441 pKa = 3.6SASAEE446 pKa = 4.06TAYY449 pKa = 10.74SSKK452 pKa = 10.83SATATYY458 pKa = 10.47INTSRR463 pKa = 11.84TSVSGVSSDD472 pKa = 3.68EE473 pKa = 3.94EE474 pKa = 4.74TVNLTTYY481 pKa = 10.05QSAYY485 pKa = 9.41AASASMVSAWSEE497 pKa = 3.97IYY499 pKa = 8.98DD500 pKa = 3.8TTISMVDD507 pKa = 3.41DD508 pKa = 3.98
MM1 pKa = 7.74ASTFAGLSIANRR13 pKa = 11.84GLTAAQVGLTVTTNNMSNIDD33 pKa = 3.34TDD35 pKa = 4.26GYY37 pKa = 10.12SRR39 pKa = 11.84QVVSQTSIGPAAVYY53 pKa = 10.45SSSLVGNGVEE63 pKa = 4.26VTSVDD68 pKa = 3.23RR69 pKa = 11.84VNSFRR74 pKa = 11.84LNQKK78 pKa = 8.46YY79 pKa = 7.33WQEE82 pKa = 3.82NSSASEE88 pKa = 4.24LEE90 pKa = 4.14VTSNYY95 pKa = 8.47MEE97 pKa = 5.26KK98 pKa = 9.87IEE100 pKa = 5.1DD101 pKa = 3.66VFGTTDD107 pKa = 2.89SSDD110 pKa = 2.92ISEE113 pKa = 4.56ALDD116 pKa = 3.34TFEE119 pKa = 6.81AEE121 pKa = 4.99LEE123 pKa = 4.27TLADD127 pKa = 4.08DD128 pKa = 4.24PTNEE132 pKa = 4.42SIRR135 pKa = 11.84KK136 pKa = 8.83IVLSYY141 pKa = 11.2AKK143 pKa = 10.18SFCDD147 pKa = 3.6TLNTAAEE154 pKa = 4.29DD155 pKa = 3.85LEE157 pKa = 4.82SIRR160 pKa = 11.84DD161 pKa = 3.71QLNTEE166 pKa = 3.92VNTAVEE172 pKa = 4.88EE173 pKa = 4.31INSYY177 pKa = 10.87ASQIAEE183 pKa = 4.49LNRR186 pKa = 11.84QISTASASGASTNEE200 pKa = 4.04LEE202 pKa = 4.45DD203 pKa = 3.51QRR205 pKa = 11.84DD206 pKa = 3.92VIVDD210 pKa = 3.65KK211 pKa = 11.37LSGLVGISVTEE222 pKa = 4.4ADD224 pKa = 4.09DD225 pKa = 3.47STYY228 pKa = 10.57TITVDD233 pKa = 3.73GVTLVKK239 pKa = 10.57GDD241 pKa = 3.73EE242 pKa = 4.17ANEE245 pKa = 4.11LEE247 pKa = 5.3CYY249 pKa = 9.12TVTDD253 pKa = 4.24SDD255 pKa = 3.87SDD257 pKa = 4.01SYY259 pKa = 11.94GMYY262 pKa = 10.22GIRR265 pKa = 11.84WAEE268 pKa = 3.87SGKK271 pKa = 10.48DD272 pKa = 3.59FDD274 pKa = 5.14SGDD277 pKa = 3.75SGSLTGYY284 pKa = 10.23LQMRR288 pKa = 11.84DD289 pKa = 3.22GSTADD294 pKa = 3.25SKK296 pKa = 11.65GVVYY300 pKa = 10.13YY301 pKa = 9.66MNQLDD306 pKa = 4.5KK307 pKa = 10.73FAQTFAQAFNEE318 pKa = 4.54GVTVTSDD325 pKa = 2.66SGTEE329 pKa = 3.9TTYY332 pKa = 10.85SGHH335 pKa = 5.85VDD337 pKa = 3.35GVGIDD342 pKa = 3.95DD343 pKa = 5.96DD344 pKa = 4.23EE345 pKa = 4.63TTGIRR350 pKa = 11.84FFTYY354 pKa = 10.77DD355 pKa = 4.0DD356 pKa = 3.6VSSADD361 pKa = 3.83FVDD364 pKa = 3.8SGSDD368 pKa = 3.57LEE370 pKa = 4.25EE371 pKa = 4.77AYY373 pKa = 11.41SNITAANISVSKK385 pKa = 10.42DD386 pKa = 3.14IQDD389 pKa = 3.84DD390 pKa = 3.96TTKK393 pKa = 10.42IAASSTAGEE402 pKa = 4.22EE403 pKa = 4.57SNTDD407 pKa = 3.48VLDD410 pKa = 4.31DD411 pKa = 5.74LIDD414 pKa = 3.58ICSDD418 pKa = 3.44VNISGNSTATEE429 pKa = 4.17MYY431 pKa = 9.55NLIVATVAGDD441 pKa = 3.6SASAEE446 pKa = 4.06TAYY449 pKa = 10.74SSKK452 pKa = 10.83SATATYY458 pKa = 10.47INTSRR463 pKa = 11.84TSVSGVSSDD472 pKa = 3.68EE473 pKa = 3.94EE474 pKa = 4.74TVNLTTYY481 pKa = 10.05QSAYY485 pKa = 9.41AASASMVSAWSEE497 pKa = 3.97IYY499 pKa = 8.98DD500 pKa = 3.8TTISMVDD507 pKa = 3.41DD508 pKa = 3.98
Molecular weight: 54.07 kDa
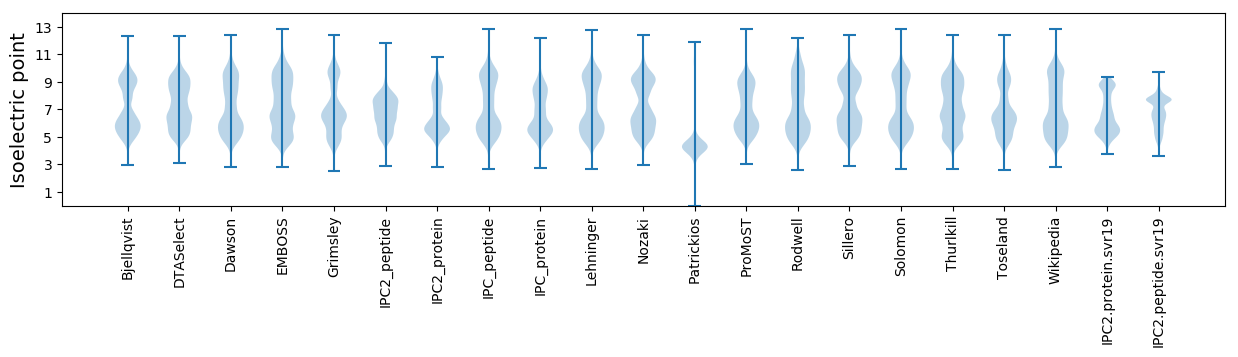
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A259V1T0|A0A259V1T0_9FIRM Amino-acid permease RocC OS=Sporomusa acidovorans DSM 3132 OX=1123286 GN=rocC_1 PE=4 SV=1
MM1 pKa = 7.8AFTRR5 pKa = 11.84SLSLSLVKK13 pKa = 10.58QGIRR17 pKa = 11.84VNGVAPGPIWTPLIPASFSDD37 pKa = 3.23RR38 pKa = 11.84HH39 pKa = 4.42VARR42 pKa = 11.84FGRR45 pKa = 11.84NTPMKK50 pKa = 10.33RR51 pKa = 11.84AGQPAEE57 pKa = 3.92VAPCYY62 pKa = 10.78VFLASQDD69 pKa = 3.57SVYY72 pKa = 10.51MSGQILHH79 pKa = 6.5VNGGTIVNGG88 pKa = 3.67
MM1 pKa = 7.8AFTRR5 pKa = 11.84SLSLSLVKK13 pKa = 10.58QGIRR17 pKa = 11.84VNGVAPGPIWTPLIPASFSDD37 pKa = 3.23RR38 pKa = 11.84HH39 pKa = 4.42VARR42 pKa = 11.84FGRR45 pKa = 11.84NTPMKK50 pKa = 10.33RR51 pKa = 11.84AGQPAEE57 pKa = 3.92VAPCYY62 pKa = 10.78VFLASQDD69 pKa = 3.57SVYY72 pKa = 10.51MSGQILHH79 pKa = 6.5VNGGTIVNGG88 pKa = 3.67
Molecular weight: 9.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1704535 |
29 |
4642 |
295.6 |
32.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.217 ± 0.04 | 1.33 ± 0.018 |
4.854 ± 0.025 | 6.104 ± 0.035 |
3.887 ± 0.022 | 7.436 ± 0.035 |
1.857 ± 0.014 | 7.433 ± 0.03 |
6.012 ± 0.029 | 9.756 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.776 ± 0.016 | 4.204 ± 0.029 |
3.961 ± 0.022 | 3.878 ± 0.023 |
4.565 ± 0.028 | 5.507 ± 0.027 |
5.54 ± 0.028 | 7.328 ± 0.029 |
1.017 ± 0.014 | 3.338 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
