
Hubei sobemo-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
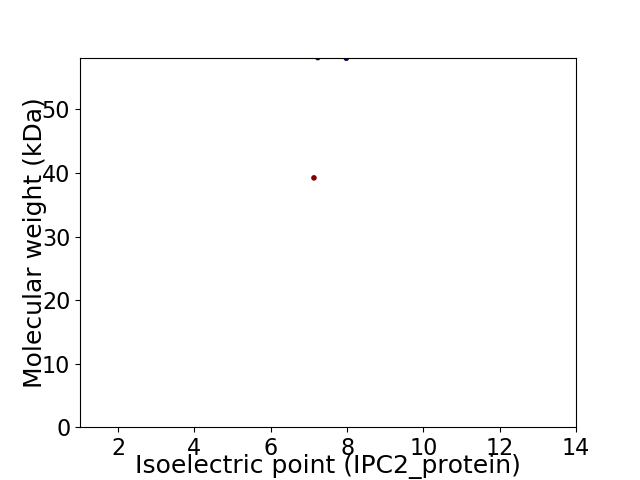
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEH3|A0A1L3KEH3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 8 OX=1923241 PE=4 SV=1
MM1 pKa = 7.53TSTPGCCILAKK12 pKa = 10.49LGSTNAQIFKK22 pKa = 9.94WDD24 pKa = 4.14GVTMDD29 pKa = 4.97PDD31 pKa = 2.9RR32 pKa = 11.84LAYY35 pKa = 9.63VRR37 pKa = 11.84AVVRR41 pKa = 11.84QRR43 pKa = 11.84TLSLLEE49 pKa = 4.43SPVADD54 pKa = 3.95PLNVFVKK61 pKa = 10.1PEE63 pKa = 3.66PHH65 pKa = 6.14SAKK68 pKa = 10.43KK69 pKa = 9.72MEE71 pKa = 4.25EE72 pKa = 3.54GRR74 pKa = 11.84YY75 pKa = 9.32RR76 pKa = 11.84LISAVSLVDD85 pKa = 3.44TLVDD89 pKa = 3.17RR90 pKa = 11.84VLFGWLGRR98 pKa = 11.84VSLDD102 pKa = 3.36SVGRR106 pKa = 11.84TPCLVGWSPISGGWRR121 pKa = 11.84QLKK124 pKa = 9.8ALYY127 pKa = 9.93RR128 pKa = 11.84GEE130 pKa = 4.24VLCLDD135 pKa = 3.85KK136 pKa = 11.31SAWDD140 pKa = 3.48WSVQGWMVDD149 pKa = 2.84MWKK152 pKa = 8.52TFVKK156 pKa = 10.16EE157 pKa = 3.65LAFGHH162 pKa = 5.83SDD164 pKa = 2.49WWGKK168 pKa = 8.79MVDD171 pKa = 3.49QRR173 pKa = 11.84FKK175 pKa = 11.5LLFEE179 pKa = 4.54DD180 pKa = 6.05AIFQFKK186 pKa = 10.84DD187 pKa = 3.19GTQVFQGSPGIMKK200 pKa = 10.02SGCYY204 pKa = 8.46LTILLNSVGQSFVHH218 pKa = 5.66YY219 pKa = 9.9LANLSLGRR227 pKa = 11.84EE228 pKa = 4.08PTQNQPHH235 pKa = 7.05TIGDD239 pKa = 3.88DD240 pKa = 3.66TVQGADD246 pKa = 3.1IDD248 pKa = 4.15FEE250 pKa = 5.22VYY252 pKa = 10.29VEE254 pKa = 4.39AIKK257 pKa = 10.88RR258 pKa = 11.84LGFRR262 pKa = 11.84VKK264 pKa = 10.23GAHH267 pKa = 5.58IRR269 pKa = 11.84KK270 pKa = 8.77NPEE273 pKa = 3.2FAGFCFGKK281 pKa = 8.3TCWPAYY287 pKa = 5.79WQKK290 pKa = 11.09HH291 pKa = 4.8LFAMAYY297 pKa = 9.7AQDD300 pKa = 4.51LGEE303 pKa = 4.33MMHH306 pKa = 8.14SYY308 pKa = 9.55MPLYY312 pKa = 10.66VNEE315 pKa = 3.91PVMFNLLRR323 pKa = 11.84RR324 pKa = 11.84VAFEE328 pKa = 4.67LNPATVLTVGEE339 pKa = 4.69CRR341 pKa = 11.84EE342 pKa = 4.16IMNGAA347 pKa = 3.56
MM1 pKa = 7.53TSTPGCCILAKK12 pKa = 10.49LGSTNAQIFKK22 pKa = 9.94WDD24 pKa = 4.14GVTMDD29 pKa = 4.97PDD31 pKa = 2.9RR32 pKa = 11.84LAYY35 pKa = 9.63VRR37 pKa = 11.84AVVRR41 pKa = 11.84QRR43 pKa = 11.84TLSLLEE49 pKa = 4.43SPVADD54 pKa = 3.95PLNVFVKK61 pKa = 10.1PEE63 pKa = 3.66PHH65 pKa = 6.14SAKK68 pKa = 10.43KK69 pKa = 9.72MEE71 pKa = 4.25EE72 pKa = 3.54GRR74 pKa = 11.84YY75 pKa = 9.32RR76 pKa = 11.84LISAVSLVDD85 pKa = 3.44TLVDD89 pKa = 3.17RR90 pKa = 11.84VLFGWLGRR98 pKa = 11.84VSLDD102 pKa = 3.36SVGRR106 pKa = 11.84TPCLVGWSPISGGWRR121 pKa = 11.84QLKK124 pKa = 9.8ALYY127 pKa = 9.93RR128 pKa = 11.84GEE130 pKa = 4.24VLCLDD135 pKa = 3.85KK136 pKa = 11.31SAWDD140 pKa = 3.48WSVQGWMVDD149 pKa = 2.84MWKK152 pKa = 8.52TFVKK156 pKa = 10.16EE157 pKa = 3.65LAFGHH162 pKa = 5.83SDD164 pKa = 2.49WWGKK168 pKa = 8.79MVDD171 pKa = 3.49QRR173 pKa = 11.84FKK175 pKa = 11.5LLFEE179 pKa = 4.54DD180 pKa = 6.05AIFQFKK186 pKa = 10.84DD187 pKa = 3.19GTQVFQGSPGIMKK200 pKa = 10.02SGCYY204 pKa = 8.46LTILLNSVGQSFVHH218 pKa = 5.66YY219 pKa = 9.9LANLSLGRR227 pKa = 11.84EE228 pKa = 4.08PTQNQPHH235 pKa = 7.05TIGDD239 pKa = 3.88DD240 pKa = 3.66TVQGADD246 pKa = 3.1IDD248 pKa = 4.15FEE250 pKa = 5.22VYY252 pKa = 10.29VEE254 pKa = 4.39AIKK257 pKa = 10.88RR258 pKa = 11.84LGFRR262 pKa = 11.84VKK264 pKa = 10.23GAHH267 pKa = 5.58IRR269 pKa = 11.84KK270 pKa = 8.77NPEE273 pKa = 3.2FAGFCFGKK281 pKa = 8.3TCWPAYY287 pKa = 5.79WQKK290 pKa = 11.09HH291 pKa = 4.8LFAMAYY297 pKa = 9.7AQDD300 pKa = 4.51LGEE303 pKa = 4.33MMHH306 pKa = 8.14SYY308 pKa = 9.55MPLYY312 pKa = 10.66VNEE315 pKa = 3.91PVMFNLLRR323 pKa = 11.84RR324 pKa = 11.84VAFEE328 pKa = 4.67LNPATVLTVGEE339 pKa = 4.69CRR341 pKa = 11.84EE342 pKa = 4.16IMNGAA347 pKa = 3.56
Molecular weight: 39.23 kDa
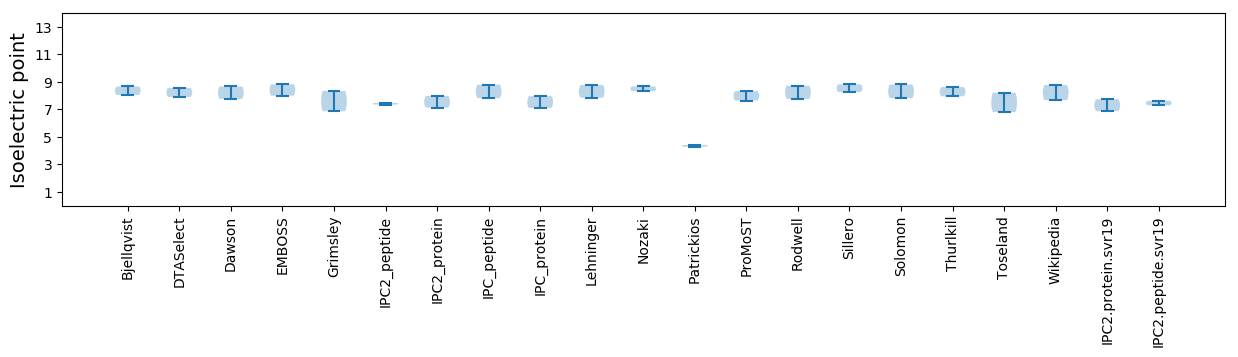
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEH3|A0A1L3KEH3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 8 OX=1923241 PE=4 SV=1
MM1 pKa = 7.49SSTMSSLISLLKK13 pKa = 10.78SFLSDD18 pKa = 2.91TCRR21 pKa = 11.84FVMTLVIMFLIWPHH35 pKa = 5.08VSEE38 pKa = 4.13WAVEE42 pKa = 3.63NWLRR46 pKa = 11.84FVDD49 pKa = 4.92LFTIPVARR57 pKa = 11.84RR58 pKa = 11.84SQAFWVHH65 pKa = 4.16QKK67 pKa = 9.21MRR69 pKa = 11.84FIGHH73 pKa = 6.04TPSDD77 pKa = 3.88HH78 pKa = 7.31VSLLEE83 pKa = 3.93IGFMAAVAYY92 pKa = 10.49CMVSTAYY99 pKa = 10.33AQISPLVKK107 pKa = 9.58VIRR110 pKa = 11.84RR111 pKa = 11.84TFVVYY116 pKa = 8.28TQQKK120 pKa = 7.24TGIYY124 pKa = 8.9SFEE127 pKa = 4.65PEE129 pKa = 3.6RR130 pKa = 11.84MMPGSSFEE138 pKa = 4.13ASSIPSFQAEE148 pKa = 4.24VIIRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.45GIYY157 pKa = 10.1YY158 pKa = 9.94QAGQGFLTSAGKK170 pKa = 10.43YY171 pKa = 9.15FVTAYY176 pKa = 10.15HH177 pKa = 5.39VVEE180 pKa = 4.55GSEE183 pKa = 4.21DD184 pKa = 3.69VQLKK188 pKa = 5.76TLKK191 pKa = 10.84GEE193 pKa = 4.46VYY195 pKa = 10.23VSPDD199 pKa = 3.42RR200 pKa = 11.84FCAMDD205 pKa = 4.25GDD207 pKa = 4.52LVYY210 pKa = 10.39MEE212 pKa = 4.83ITPQEE217 pKa = 3.9RR218 pKa = 11.84SALNLSSAKK227 pKa = 10.48LMSTAVGKK235 pKa = 10.97GSGLFVQAVAGGQKK249 pKa = 10.97SMGLLDD255 pKa = 3.2KK256 pKa = 11.13HH257 pKa = 5.83EE258 pKa = 4.4AFGYY262 pKa = 7.91VTYY265 pKa = 10.47KK266 pKa = 10.8GSTIKK271 pKa = 10.69GFSGSPYY278 pKa = 9.11YY279 pKa = 10.51VGKK282 pKa = 8.11TVYY285 pKa = 9.79GLHH288 pKa = 6.63IGGSTEE294 pKa = 3.72NLGYY298 pKa = 10.44DD299 pKa = 3.16AAYY302 pKa = 9.99ISMQLSSFEE311 pKa = 4.15EE312 pKa = 4.76DD313 pKa = 3.04SSKK316 pKa = 10.49MFIEE320 pKa = 4.07QALARR325 pKa = 11.84GDD327 pKa = 4.26EE328 pKa = 4.4INFMRR333 pKa = 11.84SPYY336 pKa = 10.41DD337 pKa = 3.24PEE339 pKa = 4.31EE340 pKa = 4.33YY341 pKa = 10.05FVEE344 pKa = 4.61HH345 pKa = 6.07NGKK348 pKa = 9.57YY349 pKa = 9.9YY350 pKa = 11.08VMEE353 pKa = 4.57EE354 pKa = 4.18EE355 pKa = 5.65DD356 pKa = 4.85LDD358 pKa = 4.18DD359 pKa = 3.9LRR361 pKa = 11.84KK362 pKa = 9.92VKK364 pKa = 10.47RR365 pKa = 11.84GSVKK369 pKa = 10.33QGILQPVYY377 pKa = 10.11EE378 pKa = 4.78RR379 pKa = 11.84EE380 pKa = 4.03SAAAGSPRR388 pKa = 11.84VDD390 pKa = 3.77EE391 pKa = 5.32DD392 pKa = 3.96LPAAPRR398 pKa = 11.84TALVFQDD405 pKa = 3.26QEE407 pKa = 4.28NLVRR411 pKa = 11.84APAVVAGARR420 pKa = 11.84GEE422 pKa = 4.17VLASQEE428 pKa = 4.07IVRR431 pKa = 11.84DD432 pKa = 3.71PLLAGPRR439 pKa = 11.84LAGLLPRR446 pKa = 11.84EE447 pKa = 4.22SLEE450 pKa = 4.37SKK452 pKa = 9.31ATAGPSQIPAVPSAPLDD469 pKa = 3.68STVKK473 pKa = 10.46SIPKK477 pKa = 8.87SARR480 pKa = 11.84ARR482 pKa = 11.84RR483 pKa = 11.84LRR485 pKa = 11.84NLRR488 pKa = 11.84QKK490 pKa = 10.07IARR493 pKa = 11.84LEE495 pKa = 4.15QPSGLTRR502 pKa = 11.84RR503 pKa = 11.84GKK505 pKa = 10.53SILDD509 pKa = 3.24RR510 pKa = 11.84RR511 pKa = 11.84VASMTFSRR519 pKa = 11.84ASSPAQQ525 pKa = 3.16
MM1 pKa = 7.49SSTMSSLISLLKK13 pKa = 10.78SFLSDD18 pKa = 2.91TCRR21 pKa = 11.84FVMTLVIMFLIWPHH35 pKa = 5.08VSEE38 pKa = 4.13WAVEE42 pKa = 3.63NWLRR46 pKa = 11.84FVDD49 pKa = 4.92LFTIPVARR57 pKa = 11.84RR58 pKa = 11.84SQAFWVHH65 pKa = 4.16QKK67 pKa = 9.21MRR69 pKa = 11.84FIGHH73 pKa = 6.04TPSDD77 pKa = 3.88HH78 pKa = 7.31VSLLEE83 pKa = 3.93IGFMAAVAYY92 pKa = 10.49CMVSTAYY99 pKa = 10.33AQISPLVKK107 pKa = 9.58VIRR110 pKa = 11.84RR111 pKa = 11.84TFVVYY116 pKa = 8.28TQQKK120 pKa = 7.24TGIYY124 pKa = 8.9SFEE127 pKa = 4.65PEE129 pKa = 3.6RR130 pKa = 11.84MMPGSSFEE138 pKa = 4.13ASSIPSFQAEE148 pKa = 4.24VIIRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.45GIYY157 pKa = 10.1YY158 pKa = 9.94QAGQGFLTSAGKK170 pKa = 10.43YY171 pKa = 9.15FVTAYY176 pKa = 10.15HH177 pKa = 5.39VVEE180 pKa = 4.55GSEE183 pKa = 4.21DD184 pKa = 3.69VQLKK188 pKa = 5.76TLKK191 pKa = 10.84GEE193 pKa = 4.46VYY195 pKa = 10.23VSPDD199 pKa = 3.42RR200 pKa = 11.84FCAMDD205 pKa = 4.25GDD207 pKa = 4.52LVYY210 pKa = 10.39MEE212 pKa = 4.83ITPQEE217 pKa = 3.9RR218 pKa = 11.84SALNLSSAKK227 pKa = 10.48LMSTAVGKK235 pKa = 10.97GSGLFVQAVAGGQKK249 pKa = 10.97SMGLLDD255 pKa = 3.2KK256 pKa = 11.13HH257 pKa = 5.83EE258 pKa = 4.4AFGYY262 pKa = 7.91VTYY265 pKa = 10.47KK266 pKa = 10.8GSTIKK271 pKa = 10.69GFSGSPYY278 pKa = 9.11YY279 pKa = 10.51VGKK282 pKa = 8.11TVYY285 pKa = 9.79GLHH288 pKa = 6.63IGGSTEE294 pKa = 3.72NLGYY298 pKa = 10.44DD299 pKa = 3.16AAYY302 pKa = 9.99ISMQLSSFEE311 pKa = 4.15EE312 pKa = 4.76DD313 pKa = 3.04SSKK316 pKa = 10.49MFIEE320 pKa = 4.07QALARR325 pKa = 11.84GDD327 pKa = 4.26EE328 pKa = 4.4INFMRR333 pKa = 11.84SPYY336 pKa = 10.41DD337 pKa = 3.24PEE339 pKa = 4.31EE340 pKa = 4.33YY341 pKa = 10.05FVEE344 pKa = 4.61HH345 pKa = 6.07NGKK348 pKa = 9.57YY349 pKa = 9.9YY350 pKa = 11.08VMEE353 pKa = 4.57EE354 pKa = 4.18EE355 pKa = 5.65DD356 pKa = 4.85LDD358 pKa = 4.18DD359 pKa = 3.9LRR361 pKa = 11.84KK362 pKa = 9.92VKK364 pKa = 10.47RR365 pKa = 11.84GSVKK369 pKa = 10.33QGILQPVYY377 pKa = 10.11EE378 pKa = 4.78RR379 pKa = 11.84EE380 pKa = 4.03SAAAGSPRR388 pKa = 11.84VDD390 pKa = 3.77EE391 pKa = 5.32DD392 pKa = 3.96LPAAPRR398 pKa = 11.84TALVFQDD405 pKa = 3.26QEE407 pKa = 4.28NLVRR411 pKa = 11.84APAVVAGARR420 pKa = 11.84GEE422 pKa = 4.17VLASQEE428 pKa = 4.07IVRR431 pKa = 11.84DD432 pKa = 3.71PLLAGPRR439 pKa = 11.84LAGLLPRR446 pKa = 11.84EE447 pKa = 4.22SLEE450 pKa = 4.37SKK452 pKa = 9.31ATAGPSQIPAVPSAPLDD469 pKa = 3.68STVKK473 pKa = 10.46SIPKK477 pKa = 8.87SARR480 pKa = 11.84ARR482 pKa = 11.84RR483 pKa = 11.84LRR485 pKa = 11.84NLRR488 pKa = 11.84QKK490 pKa = 10.07IARR493 pKa = 11.84LEE495 pKa = 4.15QPSGLTRR502 pKa = 11.84RR503 pKa = 11.84GKK505 pKa = 10.53SILDD509 pKa = 3.24RR510 pKa = 11.84RR511 pKa = 11.84VASMTFSRR519 pKa = 11.84ASSPAQQ525 pKa = 3.16
Molecular weight: 58.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872 |
347 |
525 |
436.0 |
48.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.028 ± 0.833 | 1.261 ± 0.621 |
4.702 ± 0.46 | 5.619 ± 0.6 |
4.931 ± 0.324 | 7.683 ± 0.401 |
1.72 ± 0.177 | 4.243 ± 0.467 |
4.817 ± 0.221 | 9.06 ± 0.611 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.555 ± 0.114 | 1.95 ± 0.555 |
4.931 ± 0.191 | 4.243 ± 0.124 |
6.193 ± 0.427 | 8.601 ± 1.688 |
4.472 ± 0.082 | 8.486 ± 0.266 |
1.835 ± 0.966 | 3.67 ± 0.469 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
