
Sinorhizobium fredii (strain NBRC 101917 / NGR234)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Sinorhizobium/Ensifer group; Sinorhizobium; Sinorhizobium fredii group; Sinorhizobium fredii
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
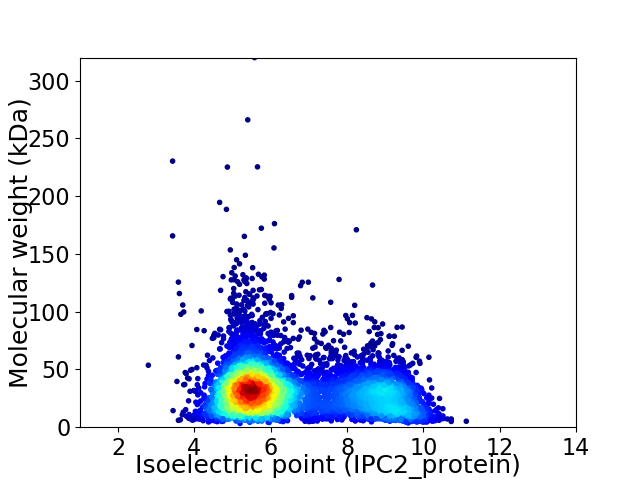
Virtual 2D-PAGE plot for 6273 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C3MB35|C3MB35_SINFN Penicillin-binding protein OS=Sinorhizobium fredii (strain NBRC 101917 / NGR234) OX=394 GN=NGR_c12470 PE=3 SV=1
MM1 pKa = 7.31GVTVSVADD9 pKa = 3.57SHH11 pKa = 6.61GLDD14 pKa = 3.15MRR16 pKa = 11.84EE17 pKa = 3.7IDD19 pKa = 4.88FSSLLYY25 pKa = 10.29ADD27 pKa = 4.09SYY29 pKa = 11.35SRR31 pKa = 11.84SSSIFSAHH39 pKa = 5.77TGNWTDD45 pKa = 3.24QFRR48 pKa = 11.84GSGFKK53 pKa = 10.53YY54 pKa = 10.4DD55 pKa = 3.51GNGYY59 pKa = 6.8PTAGTVTSYY68 pKa = 11.44AILSGGSRR76 pKa = 11.84MVLISGFSIAAAKK89 pKa = 8.17ITSAAKK95 pKa = 9.58TGSLSDD101 pKa = 3.88DD102 pKa = 3.52LAIIRR107 pKa = 11.84DD108 pKa = 3.83ALAGNDD114 pKa = 3.61NLNGGEE120 pKa = 4.02FANYY124 pKa = 9.73LRR126 pKa = 11.84GYY128 pKa = 9.7AGNDD132 pKa = 3.52VISGGQYY139 pKa = 10.6SDD141 pKa = 5.54DD142 pKa = 4.18LFGDD146 pKa = 4.26DD147 pKa = 5.45GDD149 pKa = 4.85DD150 pKa = 5.11KK151 pKa = 11.46ISGDD155 pKa = 4.04YY156 pKa = 11.04GWDD159 pKa = 3.15NLYY162 pKa = 11.04GGNGNDD168 pKa = 3.98TLNGGAGSDD177 pKa = 3.97EE178 pKa = 4.45LSGGAGSDD186 pKa = 3.23TASYY190 pKa = 10.48VGASEE195 pKa = 4.89GVVASLKK202 pKa = 10.68NLASNTNDD210 pKa = 3.12ASGDD214 pKa = 3.84SYY216 pKa = 12.02SSIEE220 pKa = 4.04NLTGTSYY227 pKa = 12.05ADD229 pKa = 3.53TLEE232 pKa = 4.45GNANANSLTGGNGNDD247 pKa = 3.41RR248 pKa = 11.84LIGRR252 pKa = 11.84AGGDD256 pKa = 3.57VLNGGAGTDD265 pKa = 3.08SAYY268 pKa = 10.95YY269 pKa = 9.67YY270 pKa = 10.38GATVGVIANLSNAAANTNDD289 pKa = 3.57AAGDD293 pKa = 3.84VYY295 pKa = 11.45SSIEE299 pKa = 4.08SLVGSSYY306 pKa = 10.27TDD308 pKa = 2.9RR309 pKa = 11.84LFGNGVANTLIGNGGHH325 pKa = 7.21DD326 pKa = 3.95SLVGYY331 pKa = 10.22AGNDD335 pKa = 3.5GLYY338 pKa = 10.42GGAGADD344 pKa = 3.42RR345 pKa = 11.84LFGGTGADD353 pKa = 2.76RR354 pKa = 11.84FIFKK358 pKa = 8.23EE359 pKa = 4.44TTEE362 pKa = 4.34STSSVTDD369 pKa = 3.7SIFDD373 pKa = 3.8FLVSEE378 pKa = 4.26QDD380 pKa = 3.53RR381 pKa = 11.84IDD383 pKa = 3.4VSGIDD388 pKa = 3.36ARR390 pKa = 11.84LSVGGDD396 pKa = 3.11QAFVFIGTAAFSGDD410 pKa = 3.38GSEE413 pKa = 4.81LRR415 pKa = 11.84YY416 pKa = 10.17EE417 pKa = 4.28RR418 pKa = 11.84LASDD422 pKa = 3.82TYY424 pKa = 10.93IYY426 pKa = 11.17ADD428 pKa = 3.26VDD430 pKa = 3.57GDD432 pKa = 3.85QVADD436 pKa = 3.83LTIHH440 pKa = 6.97LDD442 pKa = 3.59DD443 pKa = 6.55AITLTSGHH451 pKa = 6.88FILL454 pKa = 6.01
MM1 pKa = 7.31GVTVSVADD9 pKa = 3.57SHH11 pKa = 6.61GLDD14 pKa = 3.15MRR16 pKa = 11.84EE17 pKa = 3.7IDD19 pKa = 4.88FSSLLYY25 pKa = 10.29ADD27 pKa = 4.09SYY29 pKa = 11.35SRR31 pKa = 11.84SSSIFSAHH39 pKa = 5.77TGNWTDD45 pKa = 3.24QFRR48 pKa = 11.84GSGFKK53 pKa = 10.53YY54 pKa = 10.4DD55 pKa = 3.51GNGYY59 pKa = 6.8PTAGTVTSYY68 pKa = 11.44AILSGGSRR76 pKa = 11.84MVLISGFSIAAAKK89 pKa = 8.17ITSAAKK95 pKa = 9.58TGSLSDD101 pKa = 3.88DD102 pKa = 3.52LAIIRR107 pKa = 11.84DD108 pKa = 3.83ALAGNDD114 pKa = 3.61NLNGGEE120 pKa = 4.02FANYY124 pKa = 9.73LRR126 pKa = 11.84GYY128 pKa = 9.7AGNDD132 pKa = 3.52VISGGQYY139 pKa = 10.6SDD141 pKa = 5.54DD142 pKa = 4.18LFGDD146 pKa = 4.26DD147 pKa = 5.45GDD149 pKa = 4.85DD150 pKa = 5.11KK151 pKa = 11.46ISGDD155 pKa = 4.04YY156 pKa = 11.04GWDD159 pKa = 3.15NLYY162 pKa = 11.04GGNGNDD168 pKa = 3.98TLNGGAGSDD177 pKa = 3.97EE178 pKa = 4.45LSGGAGSDD186 pKa = 3.23TASYY190 pKa = 10.48VGASEE195 pKa = 4.89GVVASLKK202 pKa = 10.68NLASNTNDD210 pKa = 3.12ASGDD214 pKa = 3.84SYY216 pKa = 12.02SSIEE220 pKa = 4.04NLTGTSYY227 pKa = 12.05ADD229 pKa = 3.53TLEE232 pKa = 4.45GNANANSLTGGNGNDD247 pKa = 3.41RR248 pKa = 11.84LIGRR252 pKa = 11.84AGGDD256 pKa = 3.57VLNGGAGTDD265 pKa = 3.08SAYY268 pKa = 10.95YY269 pKa = 9.67YY270 pKa = 10.38GATVGVIANLSNAAANTNDD289 pKa = 3.57AAGDD293 pKa = 3.84VYY295 pKa = 11.45SSIEE299 pKa = 4.08SLVGSSYY306 pKa = 10.27TDD308 pKa = 2.9RR309 pKa = 11.84LFGNGVANTLIGNGGHH325 pKa = 7.21DD326 pKa = 3.95SLVGYY331 pKa = 10.22AGNDD335 pKa = 3.5GLYY338 pKa = 10.42GGAGADD344 pKa = 3.42RR345 pKa = 11.84LFGGTGADD353 pKa = 2.76RR354 pKa = 11.84FIFKK358 pKa = 8.23EE359 pKa = 4.44TTEE362 pKa = 4.34STSSVTDD369 pKa = 3.7SIFDD373 pKa = 3.8FLVSEE378 pKa = 4.26QDD380 pKa = 3.53RR381 pKa = 11.84IDD383 pKa = 3.4VSGIDD388 pKa = 3.36ARR390 pKa = 11.84LSVGGDD396 pKa = 3.11QAFVFIGTAAFSGDD410 pKa = 3.38GSEE413 pKa = 4.81LRR415 pKa = 11.84YY416 pKa = 10.17EE417 pKa = 4.28RR418 pKa = 11.84LASDD422 pKa = 3.82TYY424 pKa = 10.93IYY426 pKa = 11.17ADD428 pKa = 3.26VDD430 pKa = 3.57GDD432 pKa = 3.85QVADD436 pKa = 3.83LTIHH440 pKa = 6.97LDD442 pKa = 3.59DD443 pKa = 6.55AITLTSGHH451 pKa = 6.88FILL454 pKa = 6.01
Molecular weight: 46.4 kDa
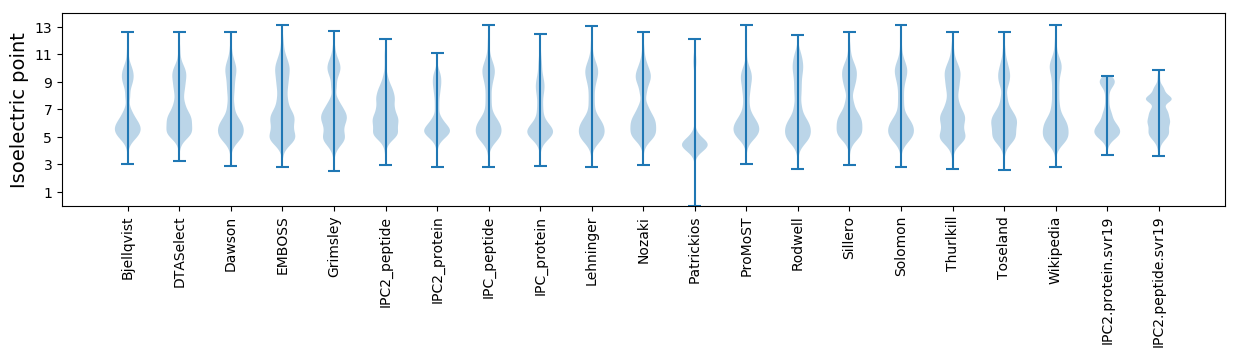
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C3MFA3|C3MFA3_SINFN Uncharacterized protein OS=Sinorhizobium fredii (strain NBRC 101917 / NGR234) OX=394 GN=NGR_c01370 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.27GGQKK29 pKa = 9.61VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.27GGQKK29 pKa = 9.61VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1950178 |
28 |
2870 |
310.9 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.144 ± 0.043 | 0.882 ± 0.01 |
5.563 ± 0.026 | 6.004 ± 0.023 |
3.861 ± 0.02 | 8.381 ± 0.027 |
2.074 ± 0.017 | 5.476 ± 0.022 |
3.542 ± 0.025 | 10.002 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.015 | 2.696 ± 0.019 |
4.937 ± 0.023 | 2.997 ± 0.02 |
7.141 ± 0.033 | 5.685 ± 0.025 |
5.201 ± 0.023 | 7.397 ± 0.027 |
1.275 ± 0.015 | 2.266 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
