
Bernardetia litoralis (strain ATCC 23117 / DSM 6794 / NBRC 15988 / NCIMB 1366 / Sio-4) (Flexibacter litoralis)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Bernardetiaceae; Bernardetia; Bernardetia litoralis
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
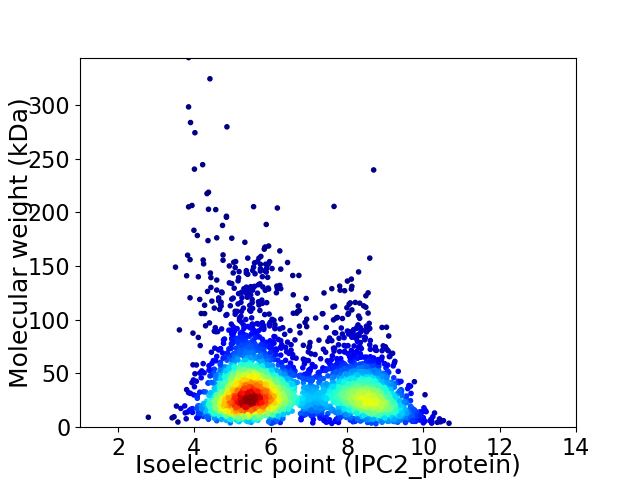
Virtual 2D-PAGE plot for 3832 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4AKN2|I4AKN2_BERLS Uncharacterized protein OS=Bernardetia litoralis (strain ATCC 23117 / DSM 6794 / NBRC 15988 / NCIMB 1366 / Sio-4) OX=880071 GN=Fleli_2136 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.42VKK4 pKa = 10.44NKK6 pKa = 8.63KK7 pKa = 6.15TTISLANYY15 pKa = 8.72CSLYY19 pKa = 9.6IDD21 pKa = 4.8KK22 pKa = 10.74FNAHH26 pKa = 5.69QLTRR30 pKa = 11.84SWQRR34 pKa = 11.84KK35 pKa = 7.2AALAAATVATSAFLGGTTSAQTIPCAPEE63 pKa = 3.56YY64 pKa = 10.8AQTASALPITNPGDD78 pKa = 3.03ITTNGGYY85 pKa = 10.76ARR87 pKa = 11.84THH89 pKa = 5.69LVDD92 pKa = 3.41YY93 pKa = 11.23DD94 pKa = 3.85GDD96 pKa = 4.03GDD98 pKa = 4.22FDD100 pKa = 5.4LMIGEE105 pKa = 4.11KK106 pKa = 9.89SGRR109 pKa = 11.84EE110 pKa = 3.38LAYY113 pKa = 10.82YY114 pKa = 9.7EE115 pKa = 4.21NTGTTSAPIWSPAVNNPNGLNLIAGEE141 pKa = 4.15NGTASFFVDD150 pKa = 3.58IDD152 pKa = 3.62NDD154 pKa = 3.66GDD156 pKa = 3.83LDD158 pKa = 4.09AFLGTEE164 pKa = 3.91KK165 pKa = 10.89DD166 pKa = 3.26GLAFFEE172 pKa = 4.4NTGTRR177 pKa = 11.84TTPVYY182 pKa = 10.1AAPVTNPFGLDD193 pKa = 3.19TGLDD197 pKa = 3.58SYY199 pKa = 9.67LTPTFADD206 pKa = 3.52IDD208 pKa = 3.85NDD210 pKa = 3.77GDD212 pKa = 3.78FDD214 pKa = 6.52LITGEE219 pKa = 4.05FGGNSRR225 pKa = 11.84FYY227 pKa = 11.12LNTGSATAPNFAAVQTNPFGLIDD250 pKa = 3.83VGFSSAPSFADD261 pKa = 3.59LDD263 pKa = 4.06GDD265 pKa = 4.09GDD267 pKa = 4.23FDD269 pKa = 6.27LMIGEE274 pKa = 5.0DD275 pKa = 3.65NGTFNYY281 pKa = 10.17FEE283 pKa = 4.86NNGTAAAAAYY293 pKa = 7.34LAPVANPFEE302 pKa = 4.9LEE304 pKa = 4.28VTSYY308 pKa = 11.0AAPTFADD315 pKa = 3.15IDD317 pKa = 4.07ADD319 pKa = 3.89GDD321 pKa = 4.02LDD323 pKa = 4.71LFAGNADD330 pKa = 3.3GDD332 pKa = 3.97AVLFTNGGSATQPDD346 pKa = 4.67FFVTPFNLPLVGDD359 pKa = 3.95YY360 pKa = 9.11TSPALADD367 pKa = 3.87LDD369 pKa = 4.02GDD371 pKa = 3.91GDD373 pKa = 3.99VDD375 pKa = 4.84VVIGDD380 pKa = 3.88SNGTITFAEE389 pKa = 4.71NIPVGGIADD398 pKa = 3.68YY399 pKa = 9.44STNSVLVNVPAGFVIPALADD419 pKa = 3.24MDD421 pKa = 4.43GDD423 pKa = 4.12GDD425 pKa = 3.6VDD427 pKa = 3.88MLVGAGDD434 pKa = 3.53GDD436 pKa = 3.96YY437 pKa = 11.2YY438 pKa = 11.29YY439 pKa = 10.64YY440 pKa = 10.81QNNGSPTASNFVLQGTNPFGLTNLGGYY467 pKa = 9.39SGPTLVDD474 pKa = 4.1FDD476 pKa = 4.41NDD478 pKa = 3.05GDD480 pKa = 3.82VDD482 pKa = 4.08VFSGTKK488 pKa = 9.75GNGVILFEE496 pKa = 4.31NTGTASAPNFAAPVFDD512 pKa = 4.25PFGIAGVGSYY522 pKa = 9.3TRR524 pKa = 11.84TTFGDD529 pKa = 3.27VDD531 pKa = 4.13SDD533 pKa = 3.71GDD535 pKa = 3.05IDD537 pKa = 3.9MFVGQKK543 pKa = 10.62DD544 pKa = 4.4GIGLTSLSYY553 pKa = 11.38YY554 pKa = 11.24NNTTGNNTQLDD565 pKa = 4.14FAPPVDD571 pKa = 4.43FPFGIQNISTWQNPALVDD589 pKa = 3.58INNDD593 pKa = 3.0GNLDD597 pKa = 3.77LFLGQKK603 pKa = 10.43DD604 pKa = 3.75DD605 pKa = 3.54GLTRR609 pKa = 11.84VYY611 pKa = 10.45TNNKK615 pKa = 8.99LFPTISIAHH624 pKa = 6.05TGNFNEE630 pKa = 4.83CNVEE634 pKa = 4.07QIEE637 pKa = 4.45ATVGNYY643 pKa = 10.39LPDD646 pKa = 4.49DD647 pKa = 4.14VTVTWYY653 pKa = 10.69NALTNLPITTGLEE666 pKa = 4.11FNPEE670 pKa = 3.97DD671 pKa = 3.5LQFRR675 pKa = 11.84ANYY678 pKa = 8.05YY679 pKa = 10.87AVVTTTDD686 pKa = 2.77GCTDD690 pKa = 3.2QSSTFSYY697 pKa = 10.94EE698 pKa = 3.84PVTPPVGTNLSAQAGYY714 pKa = 7.99NTATLDD720 pKa = 3.29WVRR723 pKa = 11.84EE724 pKa = 4.09GDD726 pKa = 3.7CDD728 pKa = 4.33SPIRR732 pKa = 11.84EE733 pKa = 4.24YY734 pKa = 10.76EE735 pKa = 4.22VYY737 pKa = 10.82ADD739 pKa = 3.74PGQFGSYY746 pKa = 10.27FLFGYY751 pKa = 10.5SSTTNFEE758 pKa = 4.07AVGLINGEE766 pKa = 4.15KK767 pKa = 9.88INFRR771 pKa = 11.84VRR773 pKa = 11.84PIFVNGTYY781 pKa = 8.9GTYY784 pKa = 11.06SNIATVKK791 pKa = 9.26PSIVLGEE798 pKa = 4.13EE799 pKa = 4.36DD800 pKa = 3.69NEE802 pKa = 4.24KK803 pKa = 10.21TGFAFFPNPNNGEE816 pKa = 4.11FNLRR820 pKa = 11.84LQDD823 pKa = 3.56GSASATVSVTSLSGQRR839 pKa = 11.84VYY841 pKa = 7.58TTRR844 pKa = 11.84MLLKK848 pKa = 10.62LLLII852 pKa = 4.83
MM1 pKa = 7.33KK2 pKa = 10.42VKK4 pKa = 10.44NKK6 pKa = 8.63KK7 pKa = 6.15TTISLANYY15 pKa = 8.72CSLYY19 pKa = 9.6IDD21 pKa = 4.8KK22 pKa = 10.74FNAHH26 pKa = 5.69QLTRR30 pKa = 11.84SWQRR34 pKa = 11.84KK35 pKa = 7.2AALAAATVATSAFLGGTTSAQTIPCAPEE63 pKa = 3.56YY64 pKa = 10.8AQTASALPITNPGDD78 pKa = 3.03ITTNGGYY85 pKa = 10.76ARR87 pKa = 11.84THH89 pKa = 5.69LVDD92 pKa = 3.41YY93 pKa = 11.23DD94 pKa = 3.85GDD96 pKa = 4.03GDD98 pKa = 4.22FDD100 pKa = 5.4LMIGEE105 pKa = 4.11KK106 pKa = 9.89SGRR109 pKa = 11.84EE110 pKa = 3.38LAYY113 pKa = 10.82YY114 pKa = 9.7EE115 pKa = 4.21NTGTTSAPIWSPAVNNPNGLNLIAGEE141 pKa = 4.15NGTASFFVDD150 pKa = 3.58IDD152 pKa = 3.62NDD154 pKa = 3.66GDD156 pKa = 3.83LDD158 pKa = 4.09AFLGTEE164 pKa = 3.91KK165 pKa = 10.89DD166 pKa = 3.26GLAFFEE172 pKa = 4.4NTGTRR177 pKa = 11.84TTPVYY182 pKa = 10.1AAPVTNPFGLDD193 pKa = 3.19TGLDD197 pKa = 3.58SYY199 pKa = 9.67LTPTFADD206 pKa = 3.52IDD208 pKa = 3.85NDD210 pKa = 3.77GDD212 pKa = 3.78FDD214 pKa = 6.52LITGEE219 pKa = 4.05FGGNSRR225 pKa = 11.84FYY227 pKa = 11.12LNTGSATAPNFAAVQTNPFGLIDD250 pKa = 3.83VGFSSAPSFADD261 pKa = 3.59LDD263 pKa = 4.06GDD265 pKa = 4.09GDD267 pKa = 4.23FDD269 pKa = 6.27LMIGEE274 pKa = 5.0DD275 pKa = 3.65NGTFNYY281 pKa = 10.17FEE283 pKa = 4.86NNGTAAAAAYY293 pKa = 7.34LAPVANPFEE302 pKa = 4.9LEE304 pKa = 4.28VTSYY308 pKa = 11.0AAPTFADD315 pKa = 3.15IDD317 pKa = 4.07ADD319 pKa = 3.89GDD321 pKa = 4.02LDD323 pKa = 4.71LFAGNADD330 pKa = 3.3GDD332 pKa = 3.97AVLFTNGGSATQPDD346 pKa = 4.67FFVTPFNLPLVGDD359 pKa = 3.95YY360 pKa = 9.11TSPALADD367 pKa = 3.87LDD369 pKa = 4.02GDD371 pKa = 3.91GDD373 pKa = 3.99VDD375 pKa = 4.84VVIGDD380 pKa = 3.88SNGTITFAEE389 pKa = 4.71NIPVGGIADD398 pKa = 3.68YY399 pKa = 9.44STNSVLVNVPAGFVIPALADD419 pKa = 3.24MDD421 pKa = 4.43GDD423 pKa = 4.12GDD425 pKa = 3.6VDD427 pKa = 3.88MLVGAGDD434 pKa = 3.53GDD436 pKa = 3.96YY437 pKa = 11.2YY438 pKa = 11.29YY439 pKa = 10.64YY440 pKa = 10.81QNNGSPTASNFVLQGTNPFGLTNLGGYY467 pKa = 9.39SGPTLVDD474 pKa = 4.1FDD476 pKa = 4.41NDD478 pKa = 3.05GDD480 pKa = 3.82VDD482 pKa = 4.08VFSGTKK488 pKa = 9.75GNGVILFEE496 pKa = 4.31NTGTASAPNFAAPVFDD512 pKa = 4.25PFGIAGVGSYY522 pKa = 9.3TRR524 pKa = 11.84TTFGDD529 pKa = 3.27VDD531 pKa = 4.13SDD533 pKa = 3.71GDD535 pKa = 3.05IDD537 pKa = 3.9MFVGQKK543 pKa = 10.62DD544 pKa = 4.4GIGLTSLSYY553 pKa = 11.38YY554 pKa = 11.24NNTTGNNTQLDD565 pKa = 4.14FAPPVDD571 pKa = 4.43FPFGIQNISTWQNPALVDD589 pKa = 3.58INNDD593 pKa = 3.0GNLDD597 pKa = 3.77LFLGQKK603 pKa = 10.43DD604 pKa = 3.75DD605 pKa = 3.54GLTRR609 pKa = 11.84VYY611 pKa = 10.45TNNKK615 pKa = 8.99LFPTISIAHH624 pKa = 6.05TGNFNEE630 pKa = 4.83CNVEE634 pKa = 4.07QIEE637 pKa = 4.45ATVGNYY643 pKa = 10.39LPDD646 pKa = 4.49DD647 pKa = 4.14VTVTWYY653 pKa = 10.69NALTNLPITTGLEE666 pKa = 4.11FNPEE670 pKa = 3.97DD671 pKa = 3.5LQFRR675 pKa = 11.84ANYY678 pKa = 8.05YY679 pKa = 10.87AVVTTTDD686 pKa = 2.77GCTDD690 pKa = 3.2QSSTFSYY697 pKa = 10.94EE698 pKa = 3.84PVTPPVGTNLSAQAGYY714 pKa = 7.99NTATLDD720 pKa = 3.29WVRR723 pKa = 11.84EE724 pKa = 4.09GDD726 pKa = 3.7CDD728 pKa = 4.33SPIRR732 pKa = 11.84EE733 pKa = 4.24YY734 pKa = 10.76EE735 pKa = 4.22VYY737 pKa = 10.82ADD739 pKa = 3.74PGQFGSYY746 pKa = 10.27FLFGYY751 pKa = 10.5SSTTNFEE758 pKa = 4.07AVGLINGEE766 pKa = 4.15KK767 pKa = 9.88INFRR771 pKa = 11.84VRR773 pKa = 11.84PIFVNGTYY781 pKa = 8.9GTYY784 pKa = 11.06SNIATVKK791 pKa = 9.26PSIVLGEE798 pKa = 4.13EE799 pKa = 4.36DD800 pKa = 3.69NEE802 pKa = 4.24KK803 pKa = 10.21TGFAFFPNPNNGEE816 pKa = 4.11FNLRR820 pKa = 11.84LQDD823 pKa = 3.56GSASATVSVTSLSGQRR839 pKa = 11.84VYY841 pKa = 7.58TTRR844 pKa = 11.84MLLKK848 pKa = 10.62LLLII852 pKa = 4.83
Molecular weight: 90.53 kDa
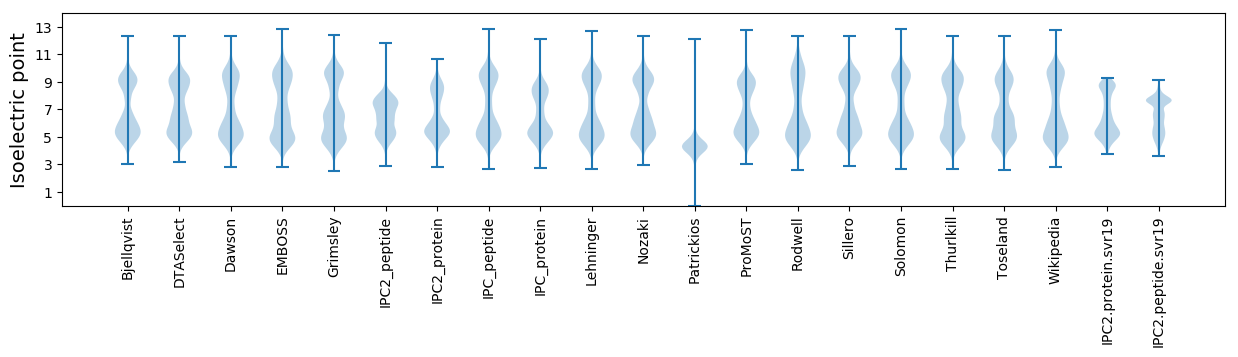
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4ALX3|I4ALX3_BERLS Membrane protein involved in the export of O-antigen and teichoic acid OS=Bernardetia litoralis (strain ATCC 23117 / DSM 6794 / NBRC 15988 / NCIMB 1366 / Sio-4) OX=880071 GN=Fleli_2595 PE=4 SV=1
MM1 pKa = 7.59AKK3 pKa = 10.09SSMIARR9 pKa = 11.84EE10 pKa = 4.01RR11 pKa = 11.84KK12 pKa = 7.3RR13 pKa = 11.84QKK15 pKa = 10.1MVDD18 pKa = 2.93KK19 pKa = 10.27YY20 pKa = 9.07ATRR23 pKa = 11.84RR24 pKa = 11.84VEE26 pKa = 4.19LKK28 pKa = 10.64KK29 pKa = 10.39LAKK32 pKa = 10.67DD33 pKa = 3.52GDD35 pKa = 4.08VEE37 pKa = 4.46AQIALDD43 pKa = 4.31KK44 pKa = 10.88LPKK47 pKa = 10.18DD48 pKa = 3.67SSPVRR53 pKa = 11.84LHH55 pKa = 5.45NRR57 pKa = 11.84CRR59 pKa = 11.84LTGRR63 pKa = 11.84PRR65 pKa = 11.84GYY67 pKa = 8.87MRR69 pKa = 11.84RR70 pKa = 11.84FGICRR75 pKa = 11.84VVFRR79 pKa = 11.84EE80 pKa = 3.73MANDD84 pKa = 3.69GKK86 pKa = 10.81IPGVTKK92 pKa = 10.74SSWW95 pKa = 2.87
MM1 pKa = 7.59AKK3 pKa = 10.09SSMIARR9 pKa = 11.84EE10 pKa = 4.01RR11 pKa = 11.84KK12 pKa = 7.3RR13 pKa = 11.84QKK15 pKa = 10.1MVDD18 pKa = 2.93KK19 pKa = 10.27YY20 pKa = 9.07ATRR23 pKa = 11.84RR24 pKa = 11.84VEE26 pKa = 4.19LKK28 pKa = 10.64KK29 pKa = 10.39LAKK32 pKa = 10.67DD33 pKa = 3.52GDD35 pKa = 4.08VEE37 pKa = 4.46AQIALDD43 pKa = 4.31KK44 pKa = 10.88LPKK47 pKa = 10.18DD48 pKa = 3.67SSPVRR53 pKa = 11.84LHH55 pKa = 5.45NRR57 pKa = 11.84CRR59 pKa = 11.84LTGRR63 pKa = 11.84PRR65 pKa = 11.84GYY67 pKa = 8.87MRR69 pKa = 11.84RR70 pKa = 11.84FGICRR75 pKa = 11.84VVFRR79 pKa = 11.84EE80 pKa = 3.73MANDD84 pKa = 3.69GKK86 pKa = 10.81IPGVTKK92 pKa = 10.74SSWW95 pKa = 2.87
Molecular weight: 10.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1339793 |
29 |
3188 |
349.6 |
39.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.819 ± 0.035 | 0.773 ± 0.014 |
5.232 ± 0.029 | 7.369 ± 0.05 |
5.522 ± 0.03 | 5.62 ± 0.041 |
1.628 ± 0.019 | 8.196 ± 0.037 |
8.191 ± 0.072 | 9.289 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.083 ± 0.019 | 6.312 ± 0.038 |
3.101 ± 0.023 | 3.92 ± 0.025 |
3.497 ± 0.024 | 6.652 ± 0.036 |
5.994 ± 0.059 | 5.57 ± 0.033 |
1.072 ± 0.012 | 4.159 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
