
Escherichia phage phiKT
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Ermolevavirus; Escherichia virus PhiKT
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
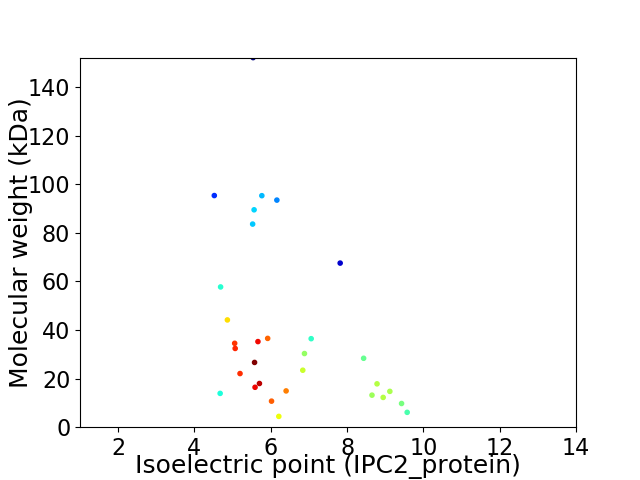
Virtual 2D-PAGE plot for 31 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H6VUC0|H6VUC0_9CAUD Uncharacterized protein OS=Escherichia phage phiKT OX=1141519 GN=phiKT_00025 PE=4 SV=1
MM1 pKa = 8.03AITFYY6 pKa = 10.48ATNNFTGDD14 pKa = 3.27GTTVNWNINFADD26 pKa = 4.97GYY28 pKa = 10.67IDD30 pKa = 3.54TTDD33 pKa = 2.79VKK35 pKa = 11.02ARR37 pKa = 11.84YY38 pKa = 9.24LDD40 pKa = 3.62NTGSYY45 pKa = 10.68VDD47 pKa = 3.62IAISSVAGNVVTISPAVADD66 pKa = 3.99GQEE69 pKa = 3.81FQIYY73 pKa = 10.01RR74 pKa = 11.84DD75 pKa = 3.64TEE77 pKa = 3.86KK78 pKa = 10.73RR79 pKa = 11.84FPLVDD84 pKa = 4.55FSDD87 pKa = 4.14GAILNEE93 pKa = 4.32TNLDD97 pKa = 3.78TLATQAVMVSAEE109 pKa = 5.25AFDD112 pKa = 3.66QSNNGVRR119 pKa = 11.84IAGDD123 pKa = 3.59SLTIAQGIDD132 pKa = 2.95AKK134 pKa = 11.17AQLALDD140 pKa = 3.7NSEE143 pKa = 4.45AAVATANGIDD153 pKa = 4.02GKK155 pKa = 10.93AQTALNTAAAAVATANAATSTAAGAVSTANTALSTANDD193 pKa = 4.0AADD196 pKa = 4.1DD197 pKa = 3.97AATAVTTANAAVTTANAATTTANAASAAASAATTTANNAANDD239 pKa = 3.57AATAVSTANTALSTANTASTNASTAVSTANAAKK272 pKa = 9.04ATAEE276 pKa = 4.5GIEE279 pKa = 4.14AKK281 pKa = 10.44ADD283 pKa = 3.45SALAASANAVTTANAAEE300 pKa = 4.13ATANAIDD307 pKa = 4.53GKK309 pKa = 10.17ATDD312 pKa = 4.46ALNTADD318 pKa = 4.59AAEE321 pKa = 4.33ATANTAAATASSAAADD337 pKa = 3.49ASTALSTVTAADD349 pKa = 3.75TKK351 pKa = 11.29AQTALDD357 pKa = 4.44NIASLDD363 pKa = 3.49AATMKK368 pKa = 10.53KK369 pKa = 10.04AANLSDD375 pKa = 3.59IADD378 pKa = 4.07KK379 pKa = 10.83AAAWLNVRR387 pKa = 11.84PIGSTPLAGDD397 pKa = 3.32PVGDD401 pKa = 3.51YY402 pKa = 11.38DD403 pKa = 4.84AVTKK407 pKa = 10.42RR408 pKa = 11.84WVEE411 pKa = 3.74NLINTGTVGPTMNGVMNYY429 pKa = 10.14GVGDD433 pKa = 3.36FHH435 pKa = 8.87LRR437 pKa = 11.84DD438 pKa = 3.4SRR440 pKa = 11.84AYY442 pKa = 7.64IQPYY446 pKa = 9.33EE447 pKa = 4.22VVSDD451 pKa = 3.9GQLLNRR457 pKa = 11.84ADD459 pKa = 3.96WPEE462 pKa = 3.82LWSYY466 pKa = 11.67AQMLSPITDD475 pKa = 4.18ADD477 pKa = 3.72WLADD481 pKa = 3.52PQKK484 pKa = 10.32RR485 pKa = 11.84GKK487 pKa = 10.55YY488 pKa = 9.69SLGNGATTFRR498 pKa = 11.84VPDD501 pKa = 3.77RR502 pKa = 11.84NGVQAGSIAALFGRR516 pKa = 11.84GDD518 pKa = 3.85GGVSSSDD525 pKa = 3.19GTILDD530 pKa = 3.74SAAPNITGSFGRR542 pKa = 11.84LVYY545 pKa = 10.79ASTGTIYY552 pKa = 10.59EE553 pKa = 4.57GNTGTGAFSAFSPQAKK569 pKa = 7.99YY570 pKa = 11.16KK571 pKa = 10.7KK572 pKa = 9.63MSDD575 pKa = 3.48LSSIDD580 pKa = 3.45AQAAEE585 pKa = 4.43YY586 pKa = 9.64PSGYY590 pKa = 10.61EE591 pKa = 3.68FFASNSSPVYY601 pKa = 10.27GRR603 pKa = 11.84GSTEE607 pKa = 3.58VRR609 pKa = 11.84PKK611 pKa = 10.88AFTGVWVIRR620 pKa = 11.84ASGGFVAANTLWSVINGDD638 pKa = 3.25ATKK641 pKa = 9.8PANNTSVNGGEE652 pKa = 4.02IQSEE656 pKa = 4.37YY657 pKa = 10.76RR658 pKa = 11.84IGTQVEE664 pKa = 4.97GKK666 pKa = 8.65TSLRR670 pKa = 11.84FVGTIGQAYY679 pKa = 8.94AARR682 pKa = 11.84LNVSNNSNTNAFIYY696 pKa = 10.83DD697 pKa = 3.33FDD699 pKa = 3.98EE700 pKa = 4.44YY701 pKa = 11.64GRR703 pKa = 11.84VNFGKK708 pKa = 9.56PGGVIKK714 pKa = 10.98SGDD717 pKa = 3.3ITLDD721 pKa = 3.19MASNGDD727 pKa = 3.81FYY729 pKa = 11.68VTTTRR734 pKa = 11.84LRR736 pKa = 11.84VEE738 pKa = 4.53HH739 pKa = 6.51PTSGGQTMGISTQGGANRR757 pKa = 11.84GADD760 pKa = 3.37QGWFEE765 pKa = 4.38HH766 pKa = 6.49WGNSYY771 pKa = 11.26GGGSDD776 pKa = 3.09KK777 pKa = 11.24SRR779 pKa = 11.84ATVVEE784 pKa = 4.08LKK786 pKa = 10.52IGTGYY791 pKa = 11.22AWYY794 pKa = 9.32SQKK797 pKa = 10.58LASGVYY803 pKa = 6.28EE804 pKa = 4.06TRR806 pKa = 11.84FASAIYY812 pKa = 8.52ATAFTQNSDD821 pKa = 3.04EE822 pKa = 4.51RR823 pKa = 11.84IKK825 pKa = 11.2EE826 pKa = 4.05NIEE829 pKa = 4.38RR830 pKa = 11.84ISDD833 pKa = 3.77PLEE836 pKa = 3.81KK837 pKa = 9.72MKK839 pKa = 10.56QIKK842 pKa = 9.23GVSWNFKK849 pKa = 8.91TVGNAKK855 pKa = 10.08GYY857 pKa = 10.78GFIAQDD863 pKa = 3.51VEE865 pKa = 4.52RR866 pKa = 11.84VLPEE870 pKa = 3.84AVSVGGGEE878 pKa = 5.14LEE880 pKa = 4.25LTDD883 pKa = 3.67GTKK886 pKa = 10.39VSDD889 pKa = 3.75VKK891 pKa = 11.11SVNTSGVAAALHH903 pKa = 6.37HH904 pKa = 5.75EE905 pKa = 5.0AILALMDD912 pKa = 3.57KK913 pKa = 11.04VEE915 pKa = 4.11ALEE918 pKa = 4.07ARR920 pKa = 11.84IAEE923 pKa = 4.42LTSQQ927 pKa = 3.58
MM1 pKa = 8.03AITFYY6 pKa = 10.48ATNNFTGDD14 pKa = 3.27GTTVNWNINFADD26 pKa = 4.97GYY28 pKa = 10.67IDD30 pKa = 3.54TTDD33 pKa = 2.79VKK35 pKa = 11.02ARR37 pKa = 11.84YY38 pKa = 9.24LDD40 pKa = 3.62NTGSYY45 pKa = 10.68VDD47 pKa = 3.62IAISSVAGNVVTISPAVADD66 pKa = 3.99GQEE69 pKa = 3.81FQIYY73 pKa = 10.01RR74 pKa = 11.84DD75 pKa = 3.64TEE77 pKa = 3.86KK78 pKa = 10.73RR79 pKa = 11.84FPLVDD84 pKa = 4.55FSDD87 pKa = 4.14GAILNEE93 pKa = 4.32TNLDD97 pKa = 3.78TLATQAVMVSAEE109 pKa = 5.25AFDD112 pKa = 3.66QSNNGVRR119 pKa = 11.84IAGDD123 pKa = 3.59SLTIAQGIDD132 pKa = 2.95AKK134 pKa = 11.17AQLALDD140 pKa = 3.7NSEE143 pKa = 4.45AAVATANGIDD153 pKa = 4.02GKK155 pKa = 10.93AQTALNTAAAAVATANAATSTAAGAVSTANTALSTANDD193 pKa = 4.0AADD196 pKa = 4.1DD197 pKa = 3.97AATAVTTANAAVTTANAATTTANAASAAASAATTTANNAANDD239 pKa = 3.57AATAVSTANTALSTANTASTNASTAVSTANAAKK272 pKa = 9.04ATAEE276 pKa = 4.5GIEE279 pKa = 4.14AKK281 pKa = 10.44ADD283 pKa = 3.45SALAASANAVTTANAAEE300 pKa = 4.13ATANAIDD307 pKa = 4.53GKK309 pKa = 10.17ATDD312 pKa = 4.46ALNTADD318 pKa = 4.59AAEE321 pKa = 4.33ATANTAAATASSAAADD337 pKa = 3.49ASTALSTVTAADD349 pKa = 3.75TKK351 pKa = 11.29AQTALDD357 pKa = 4.44NIASLDD363 pKa = 3.49AATMKK368 pKa = 10.53KK369 pKa = 10.04AANLSDD375 pKa = 3.59IADD378 pKa = 4.07KK379 pKa = 10.83AAAWLNVRR387 pKa = 11.84PIGSTPLAGDD397 pKa = 3.32PVGDD401 pKa = 3.51YY402 pKa = 11.38DD403 pKa = 4.84AVTKK407 pKa = 10.42RR408 pKa = 11.84WVEE411 pKa = 3.74NLINTGTVGPTMNGVMNYY429 pKa = 10.14GVGDD433 pKa = 3.36FHH435 pKa = 8.87LRR437 pKa = 11.84DD438 pKa = 3.4SRR440 pKa = 11.84AYY442 pKa = 7.64IQPYY446 pKa = 9.33EE447 pKa = 4.22VVSDD451 pKa = 3.9GQLLNRR457 pKa = 11.84ADD459 pKa = 3.96WPEE462 pKa = 3.82LWSYY466 pKa = 11.67AQMLSPITDD475 pKa = 4.18ADD477 pKa = 3.72WLADD481 pKa = 3.52PQKK484 pKa = 10.32RR485 pKa = 11.84GKK487 pKa = 10.55YY488 pKa = 9.69SLGNGATTFRR498 pKa = 11.84VPDD501 pKa = 3.77RR502 pKa = 11.84NGVQAGSIAALFGRR516 pKa = 11.84GDD518 pKa = 3.85GGVSSSDD525 pKa = 3.19GTILDD530 pKa = 3.74SAAPNITGSFGRR542 pKa = 11.84LVYY545 pKa = 10.79ASTGTIYY552 pKa = 10.59EE553 pKa = 4.57GNTGTGAFSAFSPQAKK569 pKa = 7.99YY570 pKa = 11.16KK571 pKa = 10.7KK572 pKa = 9.63MSDD575 pKa = 3.48LSSIDD580 pKa = 3.45AQAAEE585 pKa = 4.43YY586 pKa = 9.64PSGYY590 pKa = 10.61EE591 pKa = 3.68FFASNSSPVYY601 pKa = 10.27GRR603 pKa = 11.84GSTEE607 pKa = 3.58VRR609 pKa = 11.84PKK611 pKa = 10.88AFTGVWVIRR620 pKa = 11.84ASGGFVAANTLWSVINGDD638 pKa = 3.25ATKK641 pKa = 9.8PANNTSVNGGEE652 pKa = 4.02IQSEE656 pKa = 4.37YY657 pKa = 10.76RR658 pKa = 11.84IGTQVEE664 pKa = 4.97GKK666 pKa = 8.65TSLRR670 pKa = 11.84FVGTIGQAYY679 pKa = 8.94AARR682 pKa = 11.84LNVSNNSNTNAFIYY696 pKa = 10.83DD697 pKa = 3.33FDD699 pKa = 3.98EE700 pKa = 4.44YY701 pKa = 11.64GRR703 pKa = 11.84VNFGKK708 pKa = 9.56PGGVIKK714 pKa = 10.98SGDD717 pKa = 3.3ITLDD721 pKa = 3.19MASNGDD727 pKa = 3.81FYY729 pKa = 11.68VTTTRR734 pKa = 11.84LRR736 pKa = 11.84VEE738 pKa = 4.53HH739 pKa = 6.51PTSGGQTMGISTQGGANRR757 pKa = 11.84GADD760 pKa = 3.37QGWFEE765 pKa = 4.38HH766 pKa = 6.49WGNSYY771 pKa = 11.26GGGSDD776 pKa = 3.09KK777 pKa = 11.24SRR779 pKa = 11.84ATVVEE784 pKa = 4.08LKK786 pKa = 10.52IGTGYY791 pKa = 11.22AWYY794 pKa = 9.32SQKK797 pKa = 10.58LASGVYY803 pKa = 6.28EE804 pKa = 4.06TRR806 pKa = 11.84FASAIYY812 pKa = 8.52ATAFTQNSDD821 pKa = 3.04EE822 pKa = 4.51RR823 pKa = 11.84IKK825 pKa = 11.2EE826 pKa = 4.05NIEE829 pKa = 4.38RR830 pKa = 11.84ISDD833 pKa = 3.77PLEE836 pKa = 3.81KK837 pKa = 9.72MKK839 pKa = 10.56QIKK842 pKa = 9.23GVSWNFKK849 pKa = 8.91TVGNAKK855 pKa = 10.08GYY857 pKa = 10.78GFIAQDD863 pKa = 3.51VEE865 pKa = 4.52RR866 pKa = 11.84VLPEE870 pKa = 3.84AVSVGGGEE878 pKa = 5.14LEE880 pKa = 4.25LTDD883 pKa = 3.67GTKK886 pKa = 10.39VSDD889 pKa = 3.75VKK891 pKa = 11.11SVNTSGVAAALHH903 pKa = 6.37HH904 pKa = 5.75EE905 pKa = 5.0AILALMDD912 pKa = 3.57KK913 pKa = 11.04VEE915 pKa = 4.11ALEE918 pKa = 4.07ARR920 pKa = 11.84IAEE923 pKa = 4.42LTSQQ927 pKa = 3.58
Molecular weight: 95.38 kDa
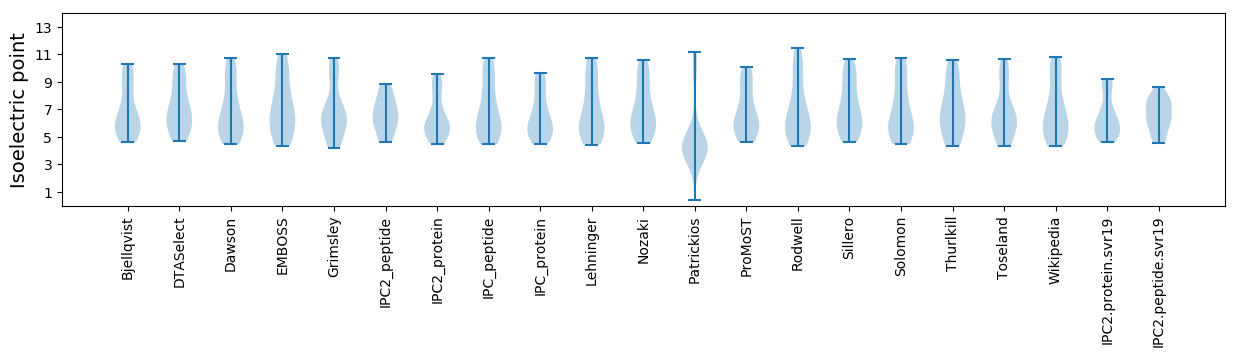
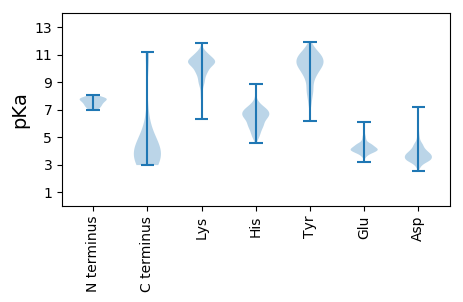
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6VUA2|H6VUA2_9CAUD Uncharacterized protein OS=Escherichia phage phiKT OX=1141519 GN=phiKT_0007 PE=4 SV=1
MM1 pKa = 7.58GEE3 pKa = 4.71CIEE6 pKa = 4.25HH7 pKa = 5.58TQVGDD12 pKa = 3.39LFGYY16 pKa = 10.89ANTRR20 pKa = 11.84RR21 pKa = 11.84GGRR24 pKa = 11.84PMKK27 pKa = 9.1MHH29 pKa = 6.86RR30 pKa = 11.84AVFLDD35 pKa = 2.89VHH37 pKa = 6.67GYY39 pKa = 8.6LPEE42 pKa = 4.32VVRR45 pKa = 11.84HH46 pKa = 5.15TCDD49 pKa = 2.85NPRR52 pKa = 11.84CINPEE57 pKa = 3.79HH58 pKa = 6.85LLAGTQKK65 pKa = 11.42DD66 pKa = 3.56NMRR69 pKa = 11.84DD70 pKa = 3.36CVEE73 pKa = 4.23RR74 pKa = 11.84GRR76 pKa = 11.84RR77 pKa = 11.84TPPPRR82 pKa = 11.84HH83 pKa = 5.89NKK85 pKa = 9.84LSQEE89 pKa = 3.9QVLEE93 pKa = 4.07IRR95 pKa = 11.84RR96 pKa = 11.84GTTPAKK102 pKa = 10.48DD103 pKa = 2.98LAEE106 pKa = 4.14RR107 pKa = 11.84FGVSKK112 pKa = 9.47VTIYY116 pKa = 10.86NVRR119 pKa = 11.84KK120 pKa = 9.79LKK122 pKa = 10.36RR123 pKa = 11.84GYY125 pKa = 9.71YY126 pKa = 9.85HH127 pKa = 7.36GG128 pKa = 4.4
MM1 pKa = 7.58GEE3 pKa = 4.71CIEE6 pKa = 4.25HH7 pKa = 5.58TQVGDD12 pKa = 3.39LFGYY16 pKa = 10.89ANTRR20 pKa = 11.84RR21 pKa = 11.84GGRR24 pKa = 11.84PMKK27 pKa = 9.1MHH29 pKa = 6.86RR30 pKa = 11.84AVFLDD35 pKa = 2.89VHH37 pKa = 6.67GYY39 pKa = 8.6LPEE42 pKa = 4.32VVRR45 pKa = 11.84HH46 pKa = 5.15TCDD49 pKa = 2.85NPRR52 pKa = 11.84CINPEE57 pKa = 3.79HH58 pKa = 6.85LLAGTQKK65 pKa = 11.42DD66 pKa = 3.56NMRR69 pKa = 11.84DD70 pKa = 3.36CVEE73 pKa = 4.23RR74 pKa = 11.84GRR76 pKa = 11.84RR77 pKa = 11.84TPPPRR82 pKa = 11.84HH83 pKa = 5.89NKK85 pKa = 9.84LSQEE89 pKa = 3.9QVLEE93 pKa = 4.07IRR95 pKa = 11.84RR96 pKa = 11.84GTTPAKK102 pKa = 10.48DD103 pKa = 2.98LAEE106 pKa = 4.14RR107 pKa = 11.84FGVSKK112 pKa = 9.47VTIYY116 pKa = 10.86NVRR119 pKa = 11.84KK120 pKa = 9.79LKK122 pKa = 10.36RR123 pKa = 11.84GYY125 pKa = 9.71YY126 pKa = 9.85HH127 pKa = 7.36GG128 pKa = 4.4
Molecular weight: 14.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11231 |
36 |
1400 |
362.3 |
39.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.275 ± 0.852 | 0.837 ± 0.189 |
6.135 ± 0.25 | 5.743 ± 0.4 |
3.428 ± 0.193 | 7.648 ± 0.331 |
1.834 ± 0.245 | 4.888 ± 0.222 |
5.529 ± 0.408 | 7.782 ± 0.333 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.992 ± 0.323 | 4.425 ± 0.288 |
3.998 ± 0.314 | 4.461 ± 0.464 |
5.218 ± 0.273 | 6.34 ± 0.366 |
6.58 ± 0.423 | 6.963 ± 0.354 |
1.416 ± 0.106 | 3.508 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
