
Rhodobacteraceae bacterium THAF1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
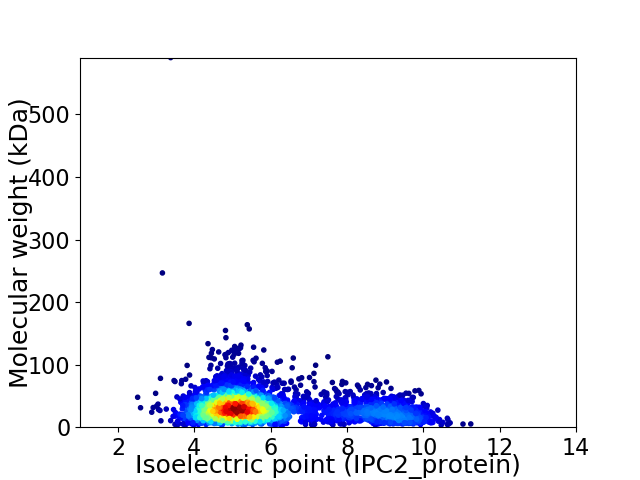
Virtual 2D-PAGE plot for 3313 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P5X3L8|A0A3P5X3L8_9RHOB Uncharacterized protein OS=Rhodobacteraceae bacterium THAF1 OX=2483814 GN=PARPLA_02764 PE=4 SV=1
MM1 pKa = 7.23MSKK4 pKa = 10.46NLLITTALCVAGLPALAQDD23 pKa = 4.04TEE25 pKa = 4.9CPVKK29 pKa = 10.93VGVLHH34 pKa = 6.32SLSGTMAISEE44 pKa = 4.52TTLKK48 pKa = 10.88DD49 pKa = 3.37AMLMLVEE56 pKa = 4.65KK57 pKa = 10.77QNADD61 pKa = 3.33GGLLGCQIEE70 pKa = 4.68TVVVDD75 pKa = 4.89PASDD79 pKa = 3.25WPRR82 pKa = 11.84FAEE85 pKa = 5.21LGRR88 pKa = 11.84QLLTEE93 pKa = 4.75DD94 pKa = 3.78EE95 pKa = 4.31VDD97 pKa = 3.86VIFGSWTSVSRR108 pKa = 11.84KK109 pKa = 9.33SVLPVLEE116 pKa = 4.27EE117 pKa = 4.15LNGLMFYY124 pKa = 10.18PVQYY128 pKa = 10.42EE129 pKa = 4.3GEE131 pKa = 4.12EE132 pKa = 3.94SSKK135 pKa = 11.08NVFYY139 pKa = 10.71TGAAPNQQAIPAVDD153 pKa = 3.57YY154 pKa = 10.58FLEE157 pKa = 4.18EE158 pKa = 4.11LGVEE162 pKa = 4.36KK163 pKa = 10.21FALLGTDD170 pKa = 3.23YY171 pKa = 11.13VYY173 pKa = 11.01PRR175 pKa = 11.84TTNNILEE182 pKa = 4.75SYY184 pKa = 10.04LQEE187 pKa = 3.84QGIAEE192 pKa = 3.95EE193 pKa = 5.25DD194 pKa = 3.28IFVNYY199 pKa = 9.64TPFGHH204 pKa = 6.47SDD206 pKa = 2.58WSTIVSDD213 pKa = 4.27VIEE216 pKa = 4.75LGSDD220 pKa = 4.15GSQVGVISTINGDD233 pKa = 3.92ANVGFYY239 pKa = 10.75KK240 pKa = 10.48EE241 pKa = 4.68LAAQDD246 pKa = 3.83VSADD250 pKa = 4.36DD251 pKa = 3.79IPVVAFSVGEE261 pKa = 4.07EE262 pKa = 4.18EE263 pKa = 5.95LSGLDD268 pKa = 3.24TSNLVGHH275 pKa = 6.85LAAWNYY281 pKa = 7.58FQSAEE286 pKa = 4.14SPEE289 pKa = 4.02NEE291 pKa = 4.11EE292 pKa = 6.12FIASWKK298 pKa = 10.3EE299 pKa = 3.73FIGDD303 pKa = 3.73EE304 pKa = 4.12NRR306 pKa = 11.84VTNDD310 pKa = 2.86PMEE313 pKa = 3.81AHH315 pKa = 6.65YY316 pKa = 10.6IGFNMWVNAVEE327 pKa = 4.26EE328 pKa = 4.46AGTTDD333 pKa = 2.78VDD335 pKa = 3.98AVRR338 pKa = 11.84EE339 pKa = 4.19AMWGQEE345 pKa = 3.93YY346 pKa = 10.11PNLTGGTAVMGTNHH360 pKa = 6.9HH361 pKa = 6.3LAKK364 pKa = 9.98PVLIGEE370 pKa = 4.31ITEE373 pKa = 5.14DD374 pKa = 3.67GQFDD378 pKa = 4.59IISEE382 pKa = 4.18TDD384 pKa = 3.79PVPGDD389 pKa = 2.9AWTDD393 pKa = 3.87FLPDD397 pKa = 3.59SAVLEE402 pKa = 4.54SDD404 pKa = 3.48WSEE407 pKa = 4.33LDD409 pKa = 3.02CGMYY413 pKa = 8.93NTEE416 pKa = 4.47TEE418 pKa = 4.25SCVQQTSNYY427 pKa = 9.55
MM1 pKa = 7.23MSKK4 pKa = 10.46NLLITTALCVAGLPALAQDD23 pKa = 4.04TEE25 pKa = 4.9CPVKK29 pKa = 10.93VGVLHH34 pKa = 6.32SLSGTMAISEE44 pKa = 4.52TTLKK48 pKa = 10.88DD49 pKa = 3.37AMLMLVEE56 pKa = 4.65KK57 pKa = 10.77QNADD61 pKa = 3.33GGLLGCQIEE70 pKa = 4.68TVVVDD75 pKa = 4.89PASDD79 pKa = 3.25WPRR82 pKa = 11.84FAEE85 pKa = 5.21LGRR88 pKa = 11.84QLLTEE93 pKa = 4.75DD94 pKa = 3.78EE95 pKa = 4.31VDD97 pKa = 3.86VIFGSWTSVSRR108 pKa = 11.84KK109 pKa = 9.33SVLPVLEE116 pKa = 4.27EE117 pKa = 4.15LNGLMFYY124 pKa = 10.18PVQYY128 pKa = 10.42EE129 pKa = 4.3GEE131 pKa = 4.12EE132 pKa = 3.94SSKK135 pKa = 11.08NVFYY139 pKa = 10.71TGAAPNQQAIPAVDD153 pKa = 3.57YY154 pKa = 10.58FLEE157 pKa = 4.18EE158 pKa = 4.11LGVEE162 pKa = 4.36KK163 pKa = 10.21FALLGTDD170 pKa = 3.23YY171 pKa = 11.13VYY173 pKa = 11.01PRR175 pKa = 11.84TTNNILEE182 pKa = 4.75SYY184 pKa = 10.04LQEE187 pKa = 3.84QGIAEE192 pKa = 3.95EE193 pKa = 5.25DD194 pKa = 3.28IFVNYY199 pKa = 9.64TPFGHH204 pKa = 6.47SDD206 pKa = 2.58WSTIVSDD213 pKa = 4.27VIEE216 pKa = 4.75LGSDD220 pKa = 4.15GSQVGVISTINGDD233 pKa = 3.92ANVGFYY239 pKa = 10.75KK240 pKa = 10.48EE241 pKa = 4.68LAAQDD246 pKa = 3.83VSADD250 pKa = 4.36DD251 pKa = 3.79IPVVAFSVGEE261 pKa = 4.07EE262 pKa = 4.18EE263 pKa = 5.95LSGLDD268 pKa = 3.24TSNLVGHH275 pKa = 6.85LAAWNYY281 pKa = 7.58FQSAEE286 pKa = 4.14SPEE289 pKa = 4.02NEE291 pKa = 4.11EE292 pKa = 6.12FIASWKK298 pKa = 10.3EE299 pKa = 3.73FIGDD303 pKa = 3.73EE304 pKa = 4.12NRR306 pKa = 11.84VTNDD310 pKa = 2.86PMEE313 pKa = 3.81AHH315 pKa = 6.65YY316 pKa = 10.6IGFNMWVNAVEE327 pKa = 4.26EE328 pKa = 4.46AGTTDD333 pKa = 2.78VDD335 pKa = 3.98AVRR338 pKa = 11.84EE339 pKa = 4.19AMWGQEE345 pKa = 3.93YY346 pKa = 10.11PNLTGGTAVMGTNHH360 pKa = 6.9HH361 pKa = 6.3LAKK364 pKa = 9.98PVLIGEE370 pKa = 4.31ITEE373 pKa = 5.14DD374 pKa = 3.67GQFDD378 pKa = 4.59IISEE382 pKa = 4.18TDD384 pKa = 3.79PVPGDD389 pKa = 2.9AWTDD393 pKa = 3.87FLPDD397 pKa = 3.59SAVLEE402 pKa = 4.54SDD404 pKa = 3.48WSEE407 pKa = 4.33LDD409 pKa = 3.02CGMYY413 pKa = 8.93NTEE416 pKa = 4.47TEE418 pKa = 4.25SCVQQTSNYY427 pKa = 9.55
Molecular weight: 46.64 kDa
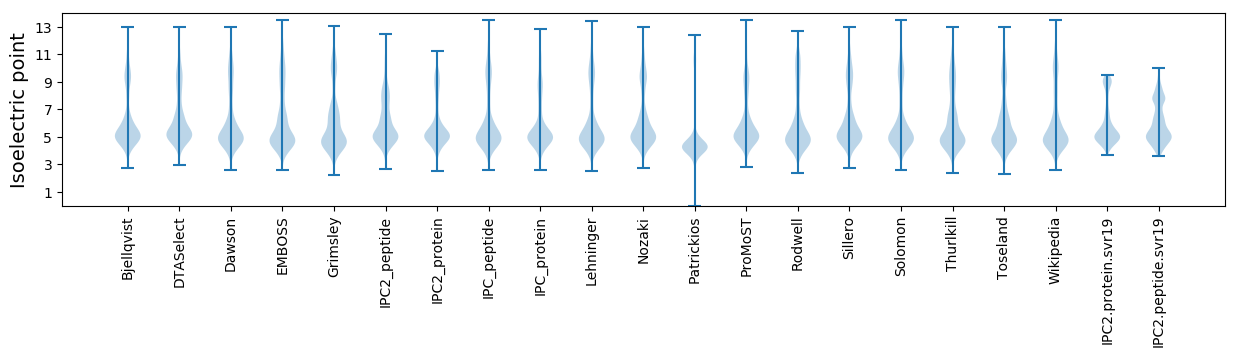
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P5X748|A0A3P5X748_9RHOB SoxAX cytochrome complex subunit A OS=Rhodobacteraceae bacterium THAF1 OX=2483814 GN=soxA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.39AGRR28 pKa = 11.84TILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.25KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.39AGRR28 pKa = 11.84TILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.25KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1009387 |
30 |
5844 |
304.7 |
32.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.498 ± 0.056 | 0.869 ± 0.013 |
6.708 ± 0.049 | 5.65 ± 0.045 |
3.603 ± 0.026 | 8.841 ± 0.049 |
1.989 ± 0.023 | 5.141 ± 0.031 |
2.775 ± 0.038 | 9.826 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.773 ± 0.027 | 2.452 ± 0.027 |
5.127 ± 0.033 | 3.145 ± 0.026 |
6.833 ± 0.059 | 5.013 ± 0.028 |
5.822 ± 0.056 | 7.392 ± 0.038 |
1.409 ± 0.019 | 2.133 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
