
Desulfomonile tiedjei (strain ATCC 49306 / DSM 6799 / DCB-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophaceae; Desulfomonile; Desulfomonile tiedjei
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
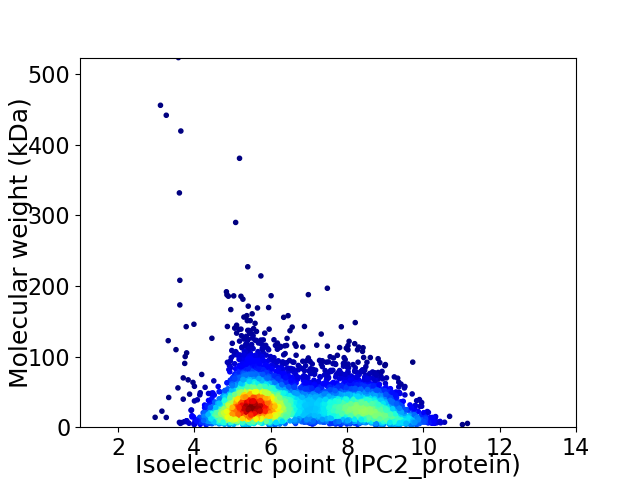
Virtual 2D-PAGE plot for 5412 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4CEW6|I4CEW6_DESTA Uncharacterized protein OS=Desulfomonile tiedjei (strain ATCC 49306 / DSM 6799 / DCB-1) OX=706587 GN=Desti_5525 PE=4 SV=1
MM1 pKa = 7.17VFANMRR7 pKa = 11.84KK8 pKa = 9.11LCRR11 pKa = 11.84EE12 pKa = 3.98LVLEE16 pKa = 4.15PLEE19 pKa = 4.45DD20 pKa = 3.55RR21 pKa = 11.84VVLDD25 pKa = 4.45AYY27 pKa = 10.98VDD29 pKa = 3.58PVYY32 pKa = 10.79QSDD35 pKa = 3.66ITVVDD40 pKa = 3.99ALAAVNHH47 pKa = 5.98EE48 pKa = 4.6PIAYY52 pKa = 8.25AQSITLVEE60 pKa = 4.23NQVSQITLTGDD71 pKa = 3.6DD72 pKa = 4.26GDD74 pKa = 4.48PDD76 pKa = 4.09VVQSLTFFLDD86 pKa = 3.51SVPANGALYY95 pKa = 10.6LSQADD100 pKa = 4.0AQNGTNPLGVGASLTTGVLWYY121 pKa = 10.46KK122 pKa = 10.62SALNNDD128 pKa = 3.31MDD130 pKa = 4.25TSFMFHH136 pKa = 6.21VKK138 pKa = 10.52DD139 pKa = 5.17DD140 pKa = 3.85GGTANGGDD148 pKa = 3.91DD149 pKa = 3.69TSASALVTVTVAPDD163 pKa = 3.61NQSPDD168 pKa = 3.12AGNFSVVVNEE178 pKa = 3.83DD179 pKa = 3.23TAIKK183 pKa = 8.21VTGWNFTDD191 pKa = 4.03AEE193 pKa = 4.64GNQGQSIRR201 pKa = 11.84ITDD204 pKa = 4.01LPDD207 pKa = 3.89HH208 pKa = 7.05GILFIDD214 pKa = 3.97ANDD217 pKa = 3.82NNVVDD222 pKa = 4.41PGEE225 pKa = 4.93AIGSDD230 pKa = 3.41QLNGRR235 pKa = 11.84VTFTFDD241 pKa = 4.48DD242 pKa = 5.27GYY244 pKa = 11.22LNNYY248 pKa = 8.63INAFPIMEE256 pKa = 4.36EE257 pKa = 4.14YY258 pKa = 10.63GVEE261 pKa = 4.2GVSLVVTGDD270 pKa = 2.69IGQNAEE276 pKa = 3.76AMTWSQLQEE285 pKa = 3.67MQAAGWEE292 pKa = 4.32IGSHH296 pKa = 5.76SMTHH300 pKa = 6.31PMLTEE305 pKa = 3.97LTDD308 pKa = 3.49SQLVYY313 pKa = 10.59EE314 pKa = 4.67LSEE317 pKa = 4.43SKK319 pKa = 10.9NLLVQHH325 pKa = 6.78GLTGTSFAYY334 pKa = 9.52PYY336 pKa = 11.07GDD338 pKa = 3.4FDD340 pKa = 4.22PRR342 pKa = 11.84VVEE345 pKa = 4.06YY346 pKa = 11.61VMDD349 pKa = 3.91YY350 pKa = 11.6YY351 pKa = 10.83EE352 pKa = 4.51GCRR355 pKa = 11.84EE356 pKa = 3.56AWGNNGINEE365 pKa = 4.54TPGNPYY371 pKa = 10.32AIYY374 pKa = 10.43NRR376 pKa = 11.84SVDD379 pKa = 3.51NTTTPQEE386 pKa = 4.15VIQWINDD393 pKa = 3.33AVANNYY399 pKa = 8.65WLVISFHH406 pKa = 6.9HH407 pKa = 6.52IVTGTPGPYY416 pKa = 9.55QYY418 pKa = 11.31NVNNFEE424 pKa = 4.58TIVSYY429 pKa = 10.8VASHH433 pKa = 6.99NIATPTISEE442 pKa = 4.27VLNNDD447 pKa = 4.24LAPISWADD455 pKa = 3.14ATTKK459 pKa = 10.5LKK461 pKa = 11.13YY462 pKa = 9.84IGSQDD467 pKa = 3.66YY468 pKa = 10.75YY469 pKa = 11.24GQDD472 pKa = 2.83SFQYY476 pKa = 10.88VVIDD480 pKa = 3.65SAGSEE485 pKa = 3.9GTMPVDD491 pKa = 3.97AGTTTVTVNPVNDD504 pKa = 4.2DD505 pKa = 4.51PINDD509 pKa = 3.66EE510 pKa = 5.13PIANPQSITLTEE522 pKa = 4.08NQVTQITLTGDD533 pKa = 3.69DD534 pKa = 4.44GDD536 pKa = 4.61PDD538 pKa = 3.72VTQTLTFFLDD548 pKa = 3.45SLPANGALYY557 pKa = 10.63LSQADD562 pKa = 3.93AQSGTNPLGVGASLSTGVLWYY583 pKa = 10.59KK584 pKa = 10.77SGLNADD590 pKa = 3.99ADD592 pKa = 4.12TSFMFHH598 pKa = 6.2VKK600 pKa = 10.73DD601 pKa = 4.18NGGTANGGDD610 pKa = 3.91DD611 pKa = 3.69TSASALVTVTVTPDD625 pKa = 3.31NQSPDD630 pKa = 3.25AGNFSTVVNEE640 pKa = 3.89DD641 pKa = 3.26TAVNVTGWNFTDD653 pKa = 4.38AEE655 pKa = 4.54GNQAQSIRR663 pKa = 11.84ITDD666 pKa = 4.02LPDD669 pKa = 3.76HH670 pKa = 6.26GTLFIDD676 pKa = 4.22ANDD679 pKa = 3.86NNVVDD684 pKa = 4.33PGEE687 pKa = 4.81AISLQPNEE695 pKa = 3.92GRR697 pKa = 11.84VTFTFDD703 pKa = 4.28DD704 pKa = 5.41GYY706 pKa = 11.63LDD708 pKa = 3.72TYY710 pKa = 10.68TNAFRR715 pKa = 11.84IMDD718 pKa = 4.0TYY720 pKa = 11.3DD721 pKa = 3.23VEE723 pKa = 4.82GVSLVVTGDD732 pKa = 3.55IGDD735 pKa = 3.6AWAMTWSQLQQMQAAGWEE753 pKa = 4.34IGSHH757 pKa = 5.76SMTHH761 pKa = 6.26PMLTGLTDD769 pKa = 3.38SQLVYY774 pKa = 10.59EE775 pKa = 4.67LSEE778 pKa = 4.48SKK780 pKa = 10.93NLLDD784 pKa = 4.44QYY786 pKa = 11.46GLTGTSFAYY795 pKa = 9.52PYY797 pKa = 11.07GDD799 pKa = 3.45FDD801 pKa = 3.98PRR803 pKa = 11.84VAEE806 pKa = 4.33FVSRR810 pKa = 11.84YY811 pKa = 9.99YY812 pKa = 10.66EE813 pKa = 4.03DD814 pKa = 5.22CRR816 pKa = 11.84EE817 pKa = 3.68AWGNNGINEE826 pKa = 4.3IPGNPYY832 pKa = 10.21AIYY835 pKa = 10.43NRR837 pKa = 11.84SVDD840 pKa = 3.51NTTTPQEE847 pKa = 4.15VIQWINDD854 pKa = 3.33AVANNYY860 pKa = 8.65WLVISFHH867 pKa = 6.84RR868 pKa = 11.84IVDD871 pKa = 4.18GTANPYY877 pKa = 9.9EE878 pKa = 4.12YY879 pKa = 10.75NVNNFEE885 pKa = 4.61TIVSYY890 pKa = 10.8VASHH894 pKa = 6.99NIATPTISEE903 pKa = 4.36VVNNDD908 pKa = 3.45LAPISWADD916 pKa = 3.14ATTKK920 pKa = 10.51LKK922 pKa = 11.13YY923 pKa = 10.28IGDD926 pKa = 3.86QEE928 pKa = 4.55YY929 pKa = 10.86SGSDD933 pKa = 2.83SFQYY937 pKa = 11.01VVIDD941 pKa = 3.66SAGSEE946 pKa = 4.11GKK948 pKa = 9.02MPADD952 pKa = 3.87AGTAIVTVIAVNDD965 pKa = 3.83APVNSIPGPQTVNGGTDD982 pKa = 3.67LLFSTANGNAISISDD997 pKa = 3.66ADD999 pKa = 3.88AGTEE1003 pKa = 3.91AVRR1006 pKa = 11.84VQLTATHH1013 pKa = 6.04GTLTLSGTSGLSFTAGDD1030 pKa = 3.95GTTDD1034 pKa = 2.76ATMTFTGSMTSINAALNGMHH1054 pKa = 6.35FTPTSNYY1061 pKa = 10.35SGDD1064 pKa = 3.4ATVQIVTNDD1073 pKa = 3.39LGNNPSGEE1081 pKa = 4.16MSDD1084 pKa = 3.48SDD1086 pKa = 4.01TVNITVVSTSGPPVAGDD1103 pKa = 3.2FAVSVNEE1110 pKa = 4.17DD1111 pKa = 2.98AVTTVSQWTFTDD1123 pKa = 3.78PNGDD1127 pKa = 3.44SAQSIRR1133 pKa = 11.84ITDD1136 pKa = 4.02LPDD1139 pKa = 3.78HH1140 pKa = 6.27GTLFLDD1146 pKa = 4.13ANTNNILDD1154 pKa = 3.95ANEE1157 pKa = 4.09AVSLNQVISWADD1169 pKa = 2.95AKK1171 pKa = 10.18TSPKK1175 pKa = 9.98VKK1177 pKa = 10.57YY1178 pKa = 10.44VGSQNYY1184 pKa = 9.53NGQDD1188 pKa = 2.99SLAYY1192 pKa = 10.19VVIDD1196 pKa = 3.36SSGEE1200 pKa = 3.77EE1201 pKa = 4.33GVAPDD1206 pKa = 3.96GDD1208 pKa = 4.16GTGSITVTAVNDD1220 pKa = 3.77APVNAVPVAQTMNANTSLIFSTANGNVISISDD1252 pKa = 3.51VDD1254 pKa = 3.88AGTDD1258 pKa = 3.43SVRR1261 pKa = 11.84VQLTATNGNLTLSGIGGLSFSAGDD1285 pKa = 3.73GTADD1289 pKa = 3.04ATMTFTGSVASINAALNGMSFTPTSNYY1316 pKa = 10.31SGAATVQIVTNDD1328 pKa = 3.45LGHH1331 pKa = 6.2NPSGEE1336 pKa = 4.1MSDD1339 pKa = 3.48SDD1341 pKa = 4.01TVNITVVSTSGPPVAGDD1358 pKa = 3.2FAVSVNEE1365 pKa = 4.82DD1366 pKa = 2.7IVATVSQWTFTDD1378 pKa = 3.78PNGDD1382 pKa = 3.44SAQSIRR1388 pKa = 11.84ITDD1391 pKa = 4.02LPDD1394 pKa = 3.78HH1395 pKa = 6.27GTLFLDD1401 pKa = 4.13ANTNNILDD1409 pKa = 3.95ANEE1412 pKa = 4.09AVSLNQVISWADD1424 pKa = 2.95AKK1426 pKa = 10.18TSPKK1430 pKa = 9.98VKK1432 pKa = 10.57YY1433 pKa = 10.44VGSQNYY1439 pKa = 9.45NGQDD1443 pKa = 2.99SLTYY1447 pKa = 10.62VVIDD1451 pKa = 3.38SSGEE1455 pKa = 3.89EE1456 pKa = 4.32GVAPAGDD1463 pKa = 3.91GTGSITVAAVNDD1475 pKa = 3.99APVNAIPAAQTMNANTSLIFSTANGNVISISDD1507 pKa = 3.51VDD1509 pKa = 4.25AGTDD1513 pKa = 3.7SISVQLTATNGNLTLAGTSGLSFTAGDD1540 pKa = 3.83GTADD1544 pKa = 3.09ATMTFTGSVASINAALNGMHH1564 pKa = 6.35FTPTSNYY1571 pKa = 9.76SGAASVQIVTNDD1583 pKa = 3.38LGHH1586 pKa = 6.2NPSGEE1591 pKa = 4.1MSDD1594 pKa = 3.48SDD1596 pKa = 4.01TVNITVVSAGGPPVADD1612 pKa = 4.71DD1613 pKa = 3.51FTLSVKK1619 pKa = 10.26EE1620 pKa = 4.04DD1621 pKa = 3.63TVVTVSKK1628 pKa = 8.47WTFTDD1633 pKa = 3.37PDD1635 pKa = 4.07GNSAQSIRR1643 pKa = 11.84ITDD1646 pKa = 4.02LPDD1649 pKa = 3.78HH1650 pKa = 6.27GTLFLDD1656 pKa = 4.12ANANNILDD1664 pKa = 3.76TSEE1667 pKa = 5.38AISLNQVISWTDD1679 pKa = 2.8AKK1681 pKa = 8.47TTPKK1685 pKa = 10.14VKK1687 pKa = 10.65YY1688 pKa = 10.29VGSQNYY1694 pKa = 9.53NGQDD1698 pKa = 2.99SLAYY1702 pKa = 10.19VVIDD1706 pKa = 3.38SSGQEE1711 pKa = 4.51GIPSAGDD1718 pKa = 3.3GTGTITVTAVNDD1730 pKa = 3.77APVNAVPGSQSINEE1744 pKa = 4.13DD1745 pKa = 3.43TVLTFSAANGNAISISDD1762 pKa = 3.54VDD1764 pKa = 4.13AGTEE1768 pKa = 3.97PVRR1771 pKa = 11.84VTLTATRR1778 pKa = 11.84GKK1780 pKa = 8.92LTLSGTTGLTFSTGDD1795 pKa = 3.37GTADD1799 pKa = 3.0TTMTFTGSIDD1809 pKa = 4.87DD1810 pKa = 4.47INAALNGMNFTPNANYY1826 pKa = 10.59AGAATVKK1833 pKa = 10.28IVTNDD1838 pKa = 3.25LGNNPSGAKK1847 pKa = 9.87SDD1849 pKa = 3.62TDD1851 pKa = 3.97TIAVTVNAIDD1861 pKa = 4.21DD1862 pKa = 4.18APVNSVPGTQTVSAGQTLVFNNVNGNRR1889 pKa = 11.84IRR1891 pKa = 11.84VSDD1894 pKa = 3.62VDD1896 pKa = 3.75AGANQIEE1903 pKa = 4.74VTLTAANGTLTLYY1916 pKa = 10.94SLTGLTFSAGDD1927 pKa = 3.69GTNDD1931 pKa = 2.7EE1932 pKa = 4.72SMTFRR1937 pKa = 11.84GTVSSINAAINRR1949 pKa = 11.84LRR1951 pKa = 11.84FIPTIAGSASLSITTNDD1968 pKa = 3.57LGASGSGGPLTDD1980 pKa = 3.6TDD1982 pKa = 4.53SIQINVTT1989 pKa = 3.25
MM1 pKa = 7.17VFANMRR7 pKa = 11.84KK8 pKa = 9.11LCRR11 pKa = 11.84EE12 pKa = 3.98LVLEE16 pKa = 4.15PLEE19 pKa = 4.45DD20 pKa = 3.55RR21 pKa = 11.84VVLDD25 pKa = 4.45AYY27 pKa = 10.98VDD29 pKa = 3.58PVYY32 pKa = 10.79QSDD35 pKa = 3.66ITVVDD40 pKa = 3.99ALAAVNHH47 pKa = 5.98EE48 pKa = 4.6PIAYY52 pKa = 8.25AQSITLVEE60 pKa = 4.23NQVSQITLTGDD71 pKa = 3.6DD72 pKa = 4.26GDD74 pKa = 4.48PDD76 pKa = 4.09VVQSLTFFLDD86 pKa = 3.51SVPANGALYY95 pKa = 10.6LSQADD100 pKa = 4.0AQNGTNPLGVGASLTTGVLWYY121 pKa = 10.46KK122 pKa = 10.62SALNNDD128 pKa = 3.31MDD130 pKa = 4.25TSFMFHH136 pKa = 6.21VKK138 pKa = 10.52DD139 pKa = 5.17DD140 pKa = 3.85GGTANGGDD148 pKa = 3.91DD149 pKa = 3.69TSASALVTVTVAPDD163 pKa = 3.61NQSPDD168 pKa = 3.12AGNFSVVVNEE178 pKa = 3.83DD179 pKa = 3.23TAIKK183 pKa = 8.21VTGWNFTDD191 pKa = 4.03AEE193 pKa = 4.64GNQGQSIRR201 pKa = 11.84ITDD204 pKa = 4.01LPDD207 pKa = 3.89HH208 pKa = 7.05GILFIDD214 pKa = 3.97ANDD217 pKa = 3.82NNVVDD222 pKa = 4.41PGEE225 pKa = 4.93AIGSDD230 pKa = 3.41QLNGRR235 pKa = 11.84VTFTFDD241 pKa = 4.48DD242 pKa = 5.27GYY244 pKa = 11.22LNNYY248 pKa = 8.63INAFPIMEE256 pKa = 4.36EE257 pKa = 4.14YY258 pKa = 10.63GVEE261 pKa = 4.2GVSLVVTGDD270 pKa = 2.69IGQNAEE276 pKa = 3.76AMTWSQLQEE285 pKa = 3.67MQAAGWEE292 pKa = 4.32IGSHH296 pKa = 5.76SMTHH300 pKa = 6.31PMLTEE305 pKa = 3.97LTDD308 pKa = 3.49SQLVYY313 pKa = 10.59EE314 pKa = 4.67LSEE317 pKa = 4.43SKK319 pKa = 10.9NLLVQHH325 pKa = 6.78GLTGTSFAYY334 pKa = 9.52PYY336 pKa = 11.07GDD338 pKa = 3.4FDD340 pKa = 4.22PRR342 pKa = 11.84VVEE345 pKa = 4.06YY346 pKa = 11.61VMDD349 pKa = 3.91YY350 pKa = 11.6YY351 pKa = 10.83EE352 pKa = 4.51GCRR355 pKa = 11.84EE356 pKa = 3.56AWGNNGINEE365 pKa = 4.54TPGNPYY371 pKa = 10.32AIYY374 pKa = 10.43NRR376 pKa = 11.84SVDD379 pKa = 3.51NTTTPQEE386 pKa = 4.15VIQWINDD393 pKa = 3.33AVANNYY399 pKa = 8.65WLVISFHH406 pKa = 6.9HH407 pKa = 6.52IVTGTPGPYY416 pKa = 9.55QYY418 pKa = 11.31NVNNFEE424 pKa = 4.58TIVSYY429 pKa = 10.8VASHH433 pKa = 6.99NIATPTISEE442 pKa = 4.27VLNNDD447 pKa = 4.24LAPISWADD455 pKa = 3.14ATTKK459 pKa = 10.5LKK461 pKa = 11.13YY462 pKa = 9.84IGSQDD467 pKa = 3.66YY468 pKa = 10.75YY469 pKa = 11.24GQDD472 pKa = 2.83SFQYY476 pKa = 10.88VVIDD480 pKa = 3.65SAGSEE485 pKa = 3.9GTMPVDD491 pKa = 3.97AGTTTVTVNPVNDD504 pKa = 4.2DD505 pKa = 4.51PINDD509 pKa = 3.66EE510 pKa = 5.13PIANPQSITLTEE522 pKa = 4.08NQVTQITLTGDD533 pKa = 3.69DD534 pKa = 4.44GDD536 pKa = 4.61PDD538 pKa = 3.72VTQTLTFFLDD548 pKa = 3.45SLPANGALYY557 pKa = 10.63LSQADD562 pKa = 3.93AQSGTNPLGVGASLSTGVLWYY583 pKa = 10.59KK584 pKa = 10.77SGLNADD590 pKa = 3.99ADD592 pKa = 4.12TSFMFHH598 pKa = 6.2VKK600 pKa = 10.73DD601 pKa = 4.18NGGTANGGDD610 pKa = 3.91DD611 pKa = 3.69TSASALVTVTVTPDD625 pKa = 3.31NQSPDD630 pKa = 3.25AGNFSTVVNEE640 pKa = 3.89DD641 pKa = 3.26TAVNVTGWNFTDD653 pKa = 4.38AEE655 pKa = 4.54GNQAQSIRR663 pKa = 11.84ITDD666 pKa = 4.02LPDD669 pKa = 3.76HH670 pKa = 6.26GTLFIDD676 pKa = 4.22ANDD679 pKa = 3.86NNVVDD684 pKa = 4.33PGEE687 pKa = 4.81AISLQPNEE695 pKa = 3.92GRR697 pKa = 11.84VTFTFDD703 pKa = 4.28DD704 pKa = 5.41GYY706 pKa = 11.63LDD708 pKa = 3.72TYY710 pKa = 10.68TNAFRR715 pKa = 11.84IMDD718 pKa = 4.0TYY720 pKa = 11.3DD721 pKa = 3.23VEE723 pKa = 4.82GVSLVVTGDD732 pKa = 3.55IGDD735 pKa = 3.6AWAMTWSQLQQMQAAGWEE753 pKa = 4.34IGSHH757 pKa = 5.76SMTHH761 pKa = 6.26PMLTGLTDD769 pKa = 3.38SQLVYY774 pKa = 10.59EE775 pKa = 4.67LSEE778 pKa = 4.48SKK780 pKa = 10.93NLLDD784 pKa = 4.44QYY786 pKa = 11.46GLTGTSFAYY795 pKa = 9.52PYY797 pKa = 11.07GDD799 pKa = 3.45FDD801 pKa = 3.98PRR803 pKa = 11.84VAEE806 pKa = 4.33FVSRR810 pKa = 11.84YY811 pKa = 9.99YY812 pKa = 10.66EE813 pKa = 4.03DD814 pKa = 5.22CRR816 pKa = 11.84EE817 pKa = 3.68AWGNNGINEE826 pKa = 4.3IPGNPYY832 pKa = 10.21AIYY835 pKa = 10.43NRR837 pKa = 11.84SVDD840 pKa = 3.51NTTTPQEE847 pKa = 4.15VIQWINDD854 pKa = 3.33AVANNYY860 pKa = 8.65WLVISFHH867 pKa = 6.84RR868 pKa = 11.84IVDD871 pKa = 4.18GTANPYY877 pKa = 9.9EE878 pKa = 4.12YY879 pKa = 10.75NVNNFEE885 pKa = 4.61TIVSYY890 pKa = 10.8VASHH894 pKa = 6.99NIATPTISEE903 pKa = 4.36VVNNDD908 pKa = 3.45LAPISWADD916 pKa = 3.14ATTKK920 pKa = 10.51LKK922 pKa = 11.13YY923 pKa = 10.28IGDD926 pKa = 3.86QEE928 pKa = 4.55YY929 pKa = 10.86SGSDD933 pKa = 2.83SFQYY937 pKa = 11.01VVIDD941 pKa = 3.66SAGSEE946 pKa = 4.11GKK948 pKa = 9.02MPADD952 pKa = 3.87AGTAIVTVIAVNDD965 pKa = 3.83APVNSIPGPQTVNGGTDD982 pKa = 3.67LLFSTANGNAISISDD997 pKa = 3.66ADD999 pKa = 3.88AGTEE1003 pKa = 3.91AVRR1006 pKa = 11.84VQLTATHH1013 pKa = 6.04GTLTLSGTSGLSFTAGDD1030 pKa = 3.95GTTDD1034 pKa = 2.76ATMTFTGSMTSINAALNGMHH1054 pKa = 6.35FTPTSNYY1061 pKa = 10.35SGDD1064 pKa = 3.4ATVQIVTNDD1073 pKa = 3.39LGNNPSGEE1081 pKa = 4.16MSDD1084 pKa = 3.48SDD1086 pKa = 4.01TVNITVVSTSGPPVAGDD1103 pKa = 3.2FAVSVNEE1110 pKa = 4.17DD1111 pKa = 2.98AVTTVSQWTFTDD1123 pKa = 3.78PNGDD1127 pKa = 3.44SAQSIRR1133 pKa = 11.84ITDD1136 pKa = 4.02LPDD1139 pKa = 3.78HH1140 pKa = 6.27GTLFLDD1146 pKa = 4.13ANTNNILDD1154 pKa = 3.95ANEE1157 pKa = 4.09AVSLNQVISWADD1169 pKa = 2.95AKK1171 pKa = 10.18TSPKK1175 pKa = 9.98VKK1177 pKa = 10.57YY1178 pKa = 10.44VGSQNYY1184 pKa = 9.53NGQDD1188 pKa = 2.99SLAYY1192 pKa = 10.19VVIDD1196 pKa = 3.36SSGEE1200 pKa = 3.77EE1201 pKa = 4.33GVAPDD1206 pKa = 3.96GDD1208 pKa = 4.16GTGSITVTAVNDD1220 pKa = 3.77APVNAVPVAQTMNANTSLIFSTANGNVISISDD1252 pKa = 3.51VDD1254 pKa = 3.88AGTDD1258 pKa = 3.43SVRR1261 pKa = 11.84VQLTATNGNLTLSGIGGLSFSAGDD1285 pKa = 3.73GTADD1289 pKa = 3.04ATMTFTGSVASINAALNGMSFTPTSNYY1316 pKa = 10.31SGAATVQIVTNDD1328 pKa = 3.45LGHH1331 pKa = 6.2NPSGEE1336 pKa = 4.1MSDD1339 pKa = 3.48SDD1341 pKa = 4.01TVNITVVSTSGPPVAGDD1358 pKa = 3.2FAVSVNEE1365 pKa = 4.82DD1366 pKa = 2.7IVATVSQWTFTDD1378 pKa = 3.78PNGDD1382 pKa = 3.44SAQSIRR1388 pKa = 11.84ITDD1391 pKa = 4.02LPDD1394 pKa = 3.78HH1395 pKa = 6.27GTLFLDD1401 pKa = 4.13ANTNNILDD1409 pKa = 3.95ANEE1412 pKa = 4.09AVSLNQVISWADD1424 pKa = 2.95AKK1426 pKa = 10.18TSPKK1430 pKa = 9.98VKK1432 pKa = 10.57YY1433 pKa = 10.44VGSQNYY1439 pKa = 9.45NGQDD1443 pKa = 2.99SLTYY1447 pKa = 10.62VVIDD1451 pKa = 3.38SSGEE1455 pKa = 3.89EE1456 pKa = 4.32GVAPAGDD1463 pKa = 3.91GTGSITVAAVNDD1475 pKa = 3.99APVNAIPAAQTMNANTSLIFSTANGNVISISDD1507 pKa = 3.51VDD1509 pKa = 4.25AGTDD1513 pKa = 3.7SISVQLTATNGNLTLAGTSGLSFTAGDD1540 pKa = 3.83GTADD1544 pKa = 3.09ATMTFTGSVASINAALNGMHH1564 pKa = 6.35FTPTSNYY1571 pKa = 9.76SGAASVQIVTNDD1583 pKa = 3.38LGHH1586 pKa = 6.2NPSGEE1591 pKa = 4.1MSDD1594 pKa = 3.48SDD1596 pKa = 4.01TVNITVVSAGGPPVADD1612 pKa = 4.71DD1613 pKa = 3.51FTLSVKK1619 pKa = 10.26EE1620 pKa = 4.04DD1621 pKa = 3.63TVVTVSKK1628 pKa = 8.47WTFTDD1633 pKa = 3.37PDD1635 pKa = 4.07GNSAQSIRR1643 pKa = 11.84ITDD1646 pKa = 4.02LPDD1649 pKa = 3.78HH1650 pKa = 6.27GTLFLDD1656 pKa = 4.12ANANNILDD1664 pKa = 3.76TSEE1667 pKa = 5.38AISLNQVISWTDD1679 pKa = 2.8AKK1681 pKa = 8.47TTPKK1685 pKa = 10.14VKK1687 pKa = 10.65YY1688 pKa = 10.29VGSQNYY1694 pKa = 9.53NGQDD1698 pKa = 2.99SLAYY1702 pKa = 10.19VVIDD1706 pKa = 3.38SSGQEE1711 pKa = 4.51GIPSAGDD1718 pKa = 3.3GTGTITVTAVNDD1730 pKa = 3.77APVNAVPGSQSINEE1744 pKa = 4.13DD1745 pKa = 3.43TVLTFSAANGNAISISDD1762 pKa = 3.54VDD1764 pKa = 4.13AGTEE1768 pKa = 3.97PVRR1771 pKa = 11.84VTLTATRR1778 pKa = 11.84GKK1780 pKa = 8.92LTLSGTTGLTFSTGDD1795 pKa = 3.37GTADD1799 pKa = 3.0TTMTFTGSIDD1809 pKa = 4.87DD1810 pKa = 4.47INAALNGMNFTPNANYY1826 pKa = 10.59AGAATVKK1833 pKa = 10.28IVTNDD1838 pKa = 3.25LGNNPSGAKK1847 pKa = 9.87SDD1849 pKa = 3.62TDD1851 pKa = 3.97TIAVTVNAIDD1861 pKa = 4.21DD1862 pKa = 4.18APVNSVPGTQTVSAGQTLVFNNVNGNRR1889 pKa = 11.84IRR1891 pKa = 11.84VSDD1894 pKa = 3.62VDD1896 pKa = 3.75AGANQIEE1903 pKa = 4.74VTLTAANGTLTLYY1916 pKa = 10.94SLTGLTFSAGDD1927 pKa = 3.69GTNDD1931 pKa = 2.7EE1932 pKa = 4.72SMTFRR1937 pKa = 11.84GTVSSINAAINRR1949 pKa = 11.84LRR1951 pKa = 11.84FIPTIAGSASLSITTNDD1968 pKa = 3.57LGASGSGGPLTDD1980 pKa = 3.6TDD1982 pKa = 4.53SIQINVTT1989 pKa = 3.25
Molecular weight: 208.11 kDa
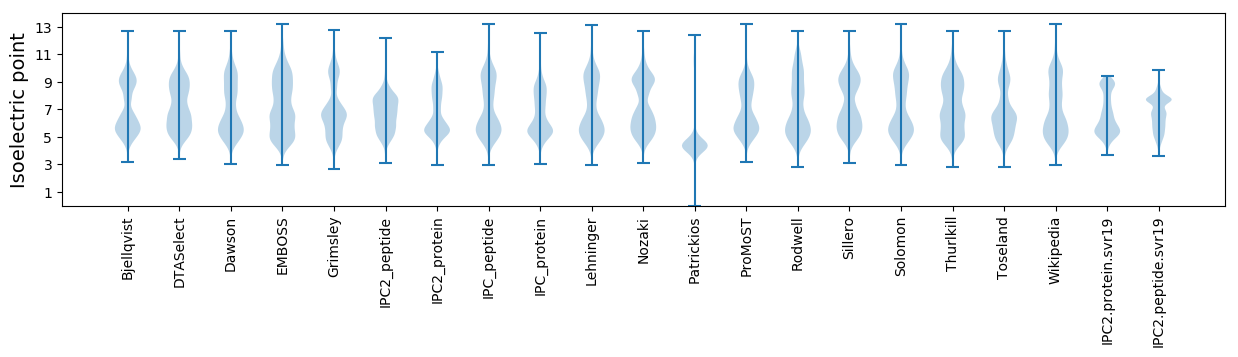
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4CAM2|I4CAM2_DESTA Uncharacterized protein OS=Desulfomonile tiedjei (strain ATCC 49306 / DSM 6799 / DCB-1) OX=706587 GN=Desti_3971 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLSRR12 pKa = 11.84KK13 pKa = 7.47RR14 pKa = 11.84THH16 pKa = 6.28GFRR19 pKa = 11.84IRR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 9.83GGRR28 pKa = 11.84LVLQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.59GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLSRR12 pKa = 11.84KK13 pKa = 7.47RR14 pKa = 11.84THH16 pKa = 6.28GFRR19 pKa = 11.84IRR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 9.83GGRR28 pKa = 11.84LVLQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.59GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1757775 |
29 |
4853 |
324.8 |
36.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.357 ± 0.038 | 1.331 ± 0.015 |
5.386 ± 0.044 | 6.611 ± 0.041 |
4.324 ± 0.025 | 7.514 ± 0.049 |
2.055 ± 0.016 | 6.456 ± 0.026 |
5.134 ± 0.04 | 10.04 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.017 | 3.395 ± 0.036 |
4.727 ± 0.027 | 3.224 ± 0.019 |
6.029 ± 0.041 | 6.309 ± 0.026 |
5.173 ± 0.047 | 7.264 ± 0.03 |
1.226 ± 0.016 | 2.827 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
