
Sphingopyxis sp. QXT-31
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis; unclassified Sphingopyxis
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
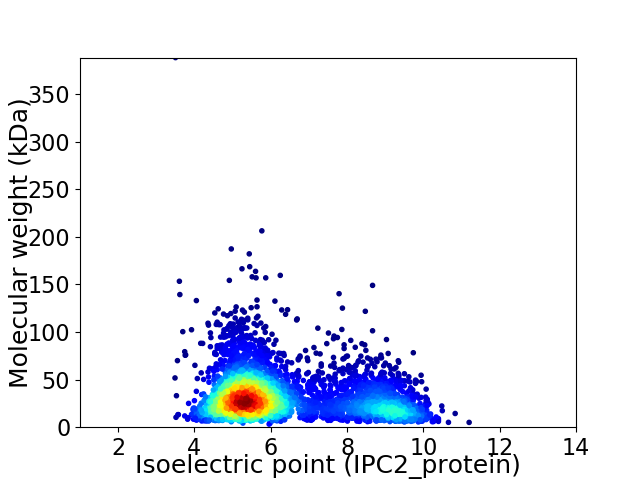
Virtual 2D-PAGE plot for 4007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
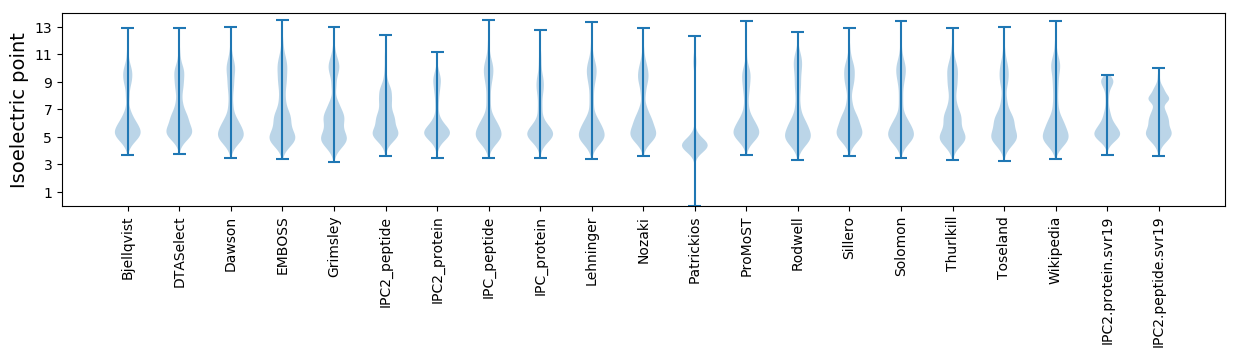
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8WW12|A0A1P8WW12_9SPHN FMN_red domain-containing protein OS=Sphingopyxis sp. QXT-31 OX=1357916 GN=BWQ93_06785 PE=4 SV=1
MM1 pKa = 7.26VLPSRR6 pKa = 11.84LQTVGEE12 pKa = 4.26SGNVFIDD19 pKa = 3.36TLVYY23 pKa = 10.39GVAFRR28 pKa = 11.84PGTTVIYY35 pKa = 10.32ALQGNPGDD43 pKa = 3.66TGLNGGAAWAANGAASAYY61 pKa = 9.93AAALASWSAVANIQFQAATVAYY83 pKa = 9.6DD84 pKa = 3.69GTGSRR89 pKa = 11.84AGYY92 pKa = 10.43DD93 pKa = 3.27FVGRR97 pKa = 11.84LSTTLPDD104 pKa = 3.72GVLGQHH110 pKa = 6.28TLPTTGDD117 pKa = 3.26MVGEE121 pKa = 4.35FNTTISYY128 pKa = 7.51FTAASNQPGGLSFLTFVHH146 pKa = 6.29EE147 pKa = 4.48LGHH150 pKa = 6.94GIGLLHH156 pKa = 6.58PHH158 pKa = 6.94ADD160 pKa = 3.72EE161 pKa = 5.56PGDD164 pKa = 4.22DD165 pKa = 4.39SFPGLNDD172 pKa = 3.33AFEE175 pKa = 4.45TGPFGLNQGIFTTMTYY191 pKa = 10.8NDD193 pKa = 4.11GYY195 pKa = 10.03EE196 pKa = 4.15DD197 pKa = 3.6VGRR200 pKa = 11.84SPSANYY206 pKa = 8.44GWQMGPGAFDD216 pKa = 3.28IAAVQYY222 pKa = 10.77LYY224 pKa = 11.14GANTTTGSGNTTYY237 pKa = 11.08TMPTLNQAGTGWVTIWDD254 pKa = 3.65VGGTDD259 pKa = 3.78TISAQGSQANAIIDD273 pKa = 3.77LRR275 pKa = 11.84AATLRR280 pKa = 11.84NEE282 pKa = 4.07EE283 pKa = 4.14GGGGFVSRR291 pKa = 11.84NGGVLGGFTIANGAVIEE308 pKa = 4.03NAIGGSGNDD317 pKa = 3.26RR318 pKa = 11.84IVGNSANNNLNGGDD332 pKa = 3.43GWDD335 pKa = 3.8TIVYY339 pKa = 9.67SHH341 pKa = 6.21MTAGVTVNLQAGTSTGDD358 pKa = 3.33GTDD361 pKa = 3.2TLVSIEE367 pKa = 4.06NAIGGNGDD375 pKa = 5.28DD376 pKa = 4.81ILTAINGRR384 pKa = 11.84DD385 pKa = 3.41IANGTQLFVKK395 pKa = 9.15TATAIISSRR404 pKa = 11.84ATAYY408 pKa = 11.1SLDD411 pKa = 3.32GLFRR415 pKa = 11.84GSGSNYY421 pKa = 9.45SVSIQAEE428 pKa = 4.35GSGGADD434 pKa = 2.8HH435 pKa = 6.91YY436 pKa = 10.77RR437 pKa = 11.84FSVANVSGPSTVTIDD452 pKa = 2.99IDD454 pKa = 3.34NSFRR458 pKa = 11.84MDD460 pKa = 4.56SIIEE464 pKa = 4.01LLDD467 pKa = 3.35SQGNVLASNDD477 pKa = 4.22DD478 pKa = 3.55GDD480 pKa = 4.62GGLDD484 pKa = 3.23PGSANIYY491 pKa = 10.69DD492 pKa = 3.83SFLTFTLPQNPATQTYY508 pKa = 8.02YY509 pKa = 10.33IRR511 pKa = 11.84VSTFSDD517 pKa = 3.42AGPASVFAGSSYY529 pKa = 9.97TLNVTANARR538 pKa = 11.84TPASGNILLGSRR550 pKa = 11.84LDD552 pKa = 4.05GGWGNDD558 pKa = 3.3QLIGGTGVDD567 pKa = 3.78TLIGGNGNDD576 pKa = 3.97TLTGGASADD585 pKa = 3.75LLYY588 pKa = 11.11GGSGDD593 pKa = 4.03DD594 pKa = 3.4NYY596 pKa = 10.53IVEE599 pKa = 4.54RR600 pKa = 11.84QDD602 pKa = 4.45DD603 pKa = 4.19LTFEE607 pKa = 4.66LLNSGVDD614 pKa = 3.41TVTASVGYY622 pKa = 9.28YY623 pKa = 10.14LYY625 pKa = 11.41AHH627 pKa = 7.14IEE629 pKa = 4.06NLTLADD635 pKa = 3.65GAGDD639 pKa = 3.37IFGVGNDD646 pKa = 4.15LNNVLTGNAGSNLMIGGLGHH666 pKa = 7.06DD667 pKa = 4.84VIRR670 pKa = 11.84GGAGVDD676 pKa = 3.38NLFGEE681 pKa = 4.53SGVDD685 pKa = 3.66QLFGDD690 pKa = 3.94AGIDD694 pKa = 3.63YY695 pKa = 10.53LVGGGGGDD703 pKa = 3.65MLDD706 pKa = 3.95GGADD710 pKa = 3.34ADD712 pKa = 3.7ALYY715 pKa = 11.03GEE717 pKa = 5.14EE718 pKa = 5.03EE719 pKa = 5.06DD720 pKa = 4.07DD721 pKa = 4.01TLIGGAGFHH730 pKa = 6.45TDD732 pKa = 2.8ILVGGTGDD740 pKa = 3.96DD741 pKa = 3.14MLYY744 pKa = 11.04ANSGLGDD751 pKa = 3.68YY752 pKa = 11.02DD753 pKa = 6.04LIDD756 pKa = 4.36GGPGNDD762 pKa = 3.32SYY764 pKa = 12.09YY765 pKa = 10.99VDD767 pKa = 4.26TPDD770 pKa = 5.16DD771 pKa = 3.83LTFEE775 pKa = 4.37ASGGGTDD782 pKa = 3.41TVYY785 pKa = 11.55ANINGAGYY793 pKa = 10.26YY794 pKa = 10.29LYY796 pKa = 11.04AHH798 pKa = 6.51TEE800 pKa = 3.94NLVLEE805 pKa = 4.77GNTPFGVGNEE815 pKa = 4.34LANQLTGNAIGNYY828 pKa = 9.52LLGGAGDD835 pKa = 4.21DD836 pKa = 4.23KK837 pKa = 11.86LNGKK841 pKa = 9.29AGNDD845 pKa = 3.37VLFGEE850 pKa = 5.42GGADD854 pKa = 2.85TFVFEE859 pKa = 5.1RR860 pKa = 11.84GTGGDD865 pKa = 3.59VIGDD869 pKa = 4.09FARR872 pKa = 11.84GTDD875 pKa = 4.31KK876 pKa = 10.89IDD878 pKa = 3.43LSAFGFANFAAVQAGFSQVGANGAINLGNGDD909 pKa = 4.95FIVLHH914 pKa = 6.05NVTMSQLAQGDD925 pKa = 4.75FILTATTPKK934 pKa = 9.98TEE936 pKa = 4.32SFGDD940 pKa = 3.8TAKK943 pKa = 10.75SAAIEE948 pKa = 4.04LADD951 pKa = 4.7AMRR954 pKa = 11.84DD955 pKa = 3.64APGAALDD962 pKa = 3.94WNPDD966 pKa = 2.38HH967 pKa = 7.47WMFHH971 pKa = 5.26YY972 pKa = 9.5QTGNNFGFAA981 pKa = 3.79
MM1 pKa = 7.26VLPSRR6 pKa = 11.84LQTVGEE12 pKa = 4.26SGNVFIDD19 pKa = 3.36TLVYY23 pKa = 10.39GVAFRR28 pKa = 11.84PGTTVIYY35 pKa = 10.32ALQGNPGDD43 pKa = 3.66TGLNGGAAWAANGAASAYY61 pKa = 9.93AAALASWSAVANIQFQAATVAYY83 pKa = 9.6DD84 pKa = 3.69GTGSRR89 pKa = 11.84AGYY92 pKa = 10.43DD93 pKa = 3.27FVGRR97 pKa = 11.84LSTTLPDD104 pKa = 3.72GVLGQHH110 pKa = 6.28TLPTTGDD117 pKa = 3.26MVGEE121 pKa = 4.35FNTTISYY128 pKa = 7.51FTAASNQPGGLSFLTFVHH146 pKa = 6.29EE147 pKa = 4.48LGHH150 pKa = 6.94GIGLLHH156 pKa = 6.58PHH158 pKa = 6.94ADD160 pKa = 3.72EE161 pKa = 5.56PGDD164 pKa = 4.22DD165 pKa = 4.39SFPGLNDD172 pKa = 3.33AFEE175 pKa = 4.45TGPFGLNQGIFTTMTYY191 pKa = 10.8NDD193 pKa = 4.11GYY195 pKa = 10.03EE196 pKa = 4.15DD197 pKa = 3.6VGRR200 pKa = 11.84SPSANYY206 pKa = 8.44GWQMGPGAFDD216 pKa = 3.28IAAVQYY222 pKa = 10.77LYY224 pKa = 11.14GANTTTGSGNTTYY237 pKa = 11.08TMPTLNQAGTGWVTIWDD254 pKa = 3.65VGGTDD259 pKa = 3.78TISAQGSQANAIIDD273 pKa = 3.77LRR275 pKa = 11.84AATLRR280 pKa = 11.84NEE282 pKa = 4.07EE283 pKa = 4.14GGGGFVSRR291 pKa = 11.84NGGVLGGFTIANGAVIEE308 pKa = 4.03NAIGGSGNDD317 pKa = 3.26RR318 pKa = 11.84IVGNSANNNLNGGDD332 pKa = 3.43GWDD335 pKa = 3.8TIVYY339 pKa = 9.67SHH341 pKa = 6.21MTAGVTVNLQAGTSTGDD358 pKa = 3.33GTDD361 pKa = 3.2TLVSIEE367 pKa = 4.06NAIGGNGDD375 pKa = 5.28DD376 pKa = 4.81ILTAINGRR384 pKa = 11.84DD385 pKa = 3.41IANGTQLFVKK395 pKa = 9.15TATAIISSRR404 pKa = 11.84ATAYY408 pKa = 11.1SLDD411 pKa = 3.32GLFRR415 pKa = 11.84GSGSNYY421 pKa = 9.45SVSIQAEE428 pKa = 4.35GSGGADD434 pKa = 2.8HH435 pKa = 6.91YY436 pKa = 10.77RR437 pKa = 11.84FSVANVSGPSTVTIDD452 pKa = 2.99IDD454 pKa = 3.34NSFRR458 pKa = 11.84MDD460 pKa = 4.56SIIEE464 pKa = 4.01LLDD467 pKa = 3.35SQGNVLASNDD477 pKa = 4.22DD478 pKa = 3.55GDD480 pKa = 4.62GGLDD484 pKa = 3.23PGSANIYY491 pKa = 10.69DD492 pKa = 3.83SFLTFTLPQNPATQTYY508 pKa = 8.02YY509 pKa = 10.33IRR511 pKa = 11.84VSTFSDD517 pKa = 3.42AGPASVFAGSSYY529 pKa = 9.97TLNVTANARR538 pKa = 11.84TPASGNILLGSRR550 pKa = 11.84LDD552 pKa = 4.05GGWGNDD558 pKa = 3.3QLIGGTGVDD567 pKa = 3.78TLIGGNGNDD576 pKa = 3.97TLTGGASADD585 pKa = 3.75LLYY588 pKa = 11.11GGSGDD593 pKa = 4.03DD594 pKa = 3.4NYY596 pKa = 10.53IVEE599 pKa = 4.54RR600 pKa = 11.84QDD602 pKa = 4.45DD603 pKa = 4.19LTFEE607 pKa = 4.66LLNSGVDD614 pKa = 3.41TVTASVGYY622 pKa = 9.28YY623 pKa = 10.14LYY625 pKa = 11.41AHH627 pKa = 7.14IEE629 pKa = 4.06NLTLADD635 pKa = 3.65GAGDD639 pKa = 3.37IFGVGNDD646 pKa = 4.15LNNVLTGNAGSNLMIGGLGHH666 pKa = 7.06DD667 pKa = 4.84VIRR670 pKa = 11.84GGAGVDD676 pKa = 3.38NLFGEE681 pKa = 4.53SGVDD685 pKa = 3.66QLFGDD690 pKa = 3.94AGIDD694 pKa = 3.63YY695 pKa = 10.53LVGGGGGDD703 pKa = 3.65MLDD706 pKa = 3.95GGADD710 pKa = 3.34ADD712 pKa = 3.7ALYY715 pKa = 11.03GEE717 pKa = 5.14EE718 pKa = 5.03EE719 pKa = 5.06DD720 pKa = 4.07DD721 pKa = 4.01TLIGGAGFHH730 pKa = 6.45TDD732 pKa = 2.8ILVGGTGDD740 pKa = 3.96DD741 pKa = 3.14MLYY744 pKa = 11.04ANSGLGDD751 pKa = 3.68YY752 pKa = 11.02DD753 pKa = 6.04LIDD756 pKa = 4.36GGPGNDD762 pKa = 3.32SYY764 pKa = 12.09YY765 pKa = 10.99VDD767 pKa = 4.26TPDD770 pKa = 5.16DD771 pKa = 3.83LTFEE775 pKa = 4.37ASGGGTDD782 pKa = 3.41TVYY785 pKa = 11.55ANINGAGYY793 pKa = 10.26YY794 pKa = 10.29LYY796 pKa = 11.04AHH798 pKa = 6.51TEE800 pKa = 3.94NLVLEE805 pKa = 4.77GNTPFGVGNEE815 pKa = 4.34LANQLTGNAIGNYY828 pKa = 9.52LLGGAGDD835 pKa = 4.21DD836 pKa = 4.23KK837 pKa = 11.86LNGKK841 pKa = 9.29AGNDD845 pKa = 3.37VLFGEE850 pKa = 5.42GGADD854 pKa = 2.85TFVFEE859 pKa = 5.1RR860 pKa = 11.84GTGGDD865 pKa = 3.59VIGDD869 pKa = 4.09FARR872 pKa = 11.84GTDD875 pKa = 4.31KK876 pKa = 10.89IDD878 pKa = 3.43LSAFGFANFAAVQAGFSQVGANGAINLGNGDD909 pKa = 4.95FIVLHH914 pKa = 6.05NVTMSQLAQGDD925 pKa = 4.75FILTATTPKK934 pKa = 9.98TEE936 pKa = 4.32SFGDD940 pKa = 3.8TAKK943 pKa = 10.75SAAIEE948 pKa = 4.04LADD951 pKa = 4.7AMRR954 pKa = 11.84DD955 pKa = 3.64APGAALDD962 pKa = 3.94WNPDD966 pKa = 2.38HH967 pKa = 7.47WMFHH971 pKa = 5.26YY972 pKa = 9.5QTGNNFGFAA981 pKa = 3.79
Molecular weight: 100.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8X191|A0A1P8X191_9SPHN Proprotein convertase P OS=Sphingopyxis sp. QXT-31 OX=1357916 GN=BWQ93_17610 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.43GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.1VLANRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.26KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.43GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.1VLANRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.26KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276034 |
29 |
4048 |
318.5 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.941 ± 0.061 | 0.74 ± 0.011 |
6.201 ± 0.034 | 5.359 ± 0.035 |
3.575 ± 0.023 | 9.198 ± 0.046 |
1.935 ± 0.021 | 5.027 ± 0.024 |
3.137 ± 0.035 | 9.716 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.391 ± 0.019 | 2.528 ± 0.031 |
5.369 ± 0.03 | 2.892 ± 0.023 |
6.999 ± 0.039 | 5.024 ± 0.028 |
5.181 ± 0.042 | 7.037 ± 0.032 |
1.499 ± 0.016 | 2.252 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
