
Alkalitalea saponilacus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinilabiliaceae; Alkalitalea
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
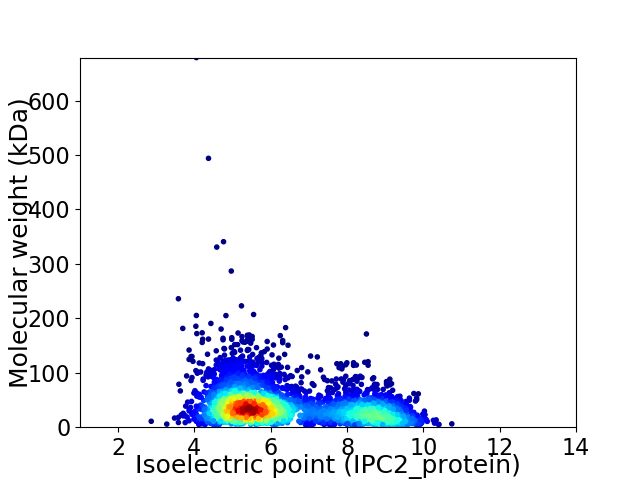
Virtual 2D-PAGE plot for 3564 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T5HMG7|A0A1T5HMG7_9BACT Gliding motility-associated protein GldL OS=Alkalitalea saponilacus OX=889453 GN=SAMN03080601_02461 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.65YY3 pKa = 10.05KK4 pKa = 10.45VLSIALLTFVLTSCYY19 pKa = 10.1DD20 pKa = 3.14RR21 pKa = 11.84EE22 pKa = 4.2YY23 pKa = 10.77FQLAEE28 pKa = 4.21TVQPVLSSSEE38 pKa = 4.4SISDD42 pKa = 3.62VVIEE46 pKa = 5.07EE47 pKa = 4.64DD48 pKa = 4.8ALDD51 pKa = 3.5QNFTVFSWTAASFGDD66 pKa = 3.85GLPDD70 pKa = 3.19AEE72 pKa = 4.49YY73 pKa = 11.2ALRR76 pKa = 11.84MDD78 pKa = 4.4LAGNNFQNPISLGRR92 pKa = 11.84TKK94 pKa = 10.66SLSISVLNSRR104 pKa = 11.84MNQNLLSIGIEE115 pKa = 3.97PGNPASIEE123 pKa = 4.21FQLHH127 pKa = 5.89ASINPDD133 pKa = 2.45VSTISNIVTSTVTPLEE149 pKa = 4.08VVIEE153 pKa = 4.16YY154 pKa = 7.44PTIYY158 pKa = 10.53LAGDD162 pKa = 3.4HH163 pKa = 6.54NGWSFNDD170 pKa = 3.13QLYY173 pKa = 11.32SMRR176 pKa = 11.84SDD178 pKa = 3.9EE179 pKa = 4.23IYY181 pKa = 10.89EE182 pKa = 4.1GFIYY186 pKa = 9.78MDD188 pKa = 4.9NGDD191 pKa = 3.19NWIGFKK197 pKa = 9.91MSYY200 pKa = 9.54EE201 pKa = 4.09PDD203 pKa = 3.48WIEE206 pKa = 4.58DD207 pKa = 3.86LVIGDD212 pKa = 4.85PDD214 pKa = 3.85DD215 pKa = 4.82SGTSGTLQVGNWGGSNIYY233 pKa = 8.85ATEE236 pKa = 4.26GPGYY240 pKa = 10.43YY241 pKa = 9.73RR242 pKa = 11.84IRR244 pKa = 11.84ANLVNNTISILKK256 pKa = 8.09TEE258 pKa = 4.07WAITGDD264 pKa = 3.61FNGWSFTPLVYY275 pKa = 9.83DD276 pKa = 3.69TDD278 pKa = 4.17DD279 pKa = 4.12KK280 pKa = 11.57VWSATIDD287 pKa = 3.34MTAGGFKK294 pKa = 10.56FIANEE299 pKa = 3.73DD300 pKa = 3.22WSLVFGDD307 pKa = 4.69DD308 pKa = 4.61DD309 pKa = 5.51LNGVLDD315 pKa = 4.95PGRR318 pKa = 11.84DD319 pKa = 3.64DD320 pKa = 4.75NNIQITEE327 pKa = 4.32DD328 pKa = 3.32GNYY331 pKa = 9.28TIIMDD336 pKa = 4.49LSGAPYY342 pKa = 10.12VYY344 pKa = 10.53SIVKK348 pKa = 9.73NN349 pKa = 3.77
MM1 pKa = 7.69KK2 pKa = 10.65YY3 pKa = 10.05KK4 pKa = 10.45VLSIALLTFVLTSCYY19 pKa = 10.1DD20 pKa = 3.14RR21 pKa = 11.84EE22 pKa = 4.2YY23 pKa = 10.77FQLAEE28 pKa = 4.21TVQPVLSSSEE38 pKa = 4.4SISDD42 pKa = 3.62VVIEE46 pKa = 5.07EE47 pKa = 4.64DD48 pKa = 4.8ALDD51 pKa = 3.5QNFTVFSWTAASFGDD66 pKa = 3.85GLPDD70 pKa = 3.19AEE72 pKa = 4.49YY73 pKa = 11.2ALRR76 pKa = 11.84MDD78 pKa = 4.4LAGNNFQNPISLGRR92 pKa = 11.84TKK94 pKa = 10.66SLSISVLNSRR104 pKa = 11.84MNQNLLSIGIEE115 pKa = 3.97PGNPASIEE123 pKa = 4.21FQLHH127 pKa = 5.89ASINPDD133 pKa = 2.45VSTISNIVTSTVTPLEE149 pKa = 4.08VVIEE153 pKa = 4.16YY154 pKa = 7.44PTIYY158 pKa = 10.53LAGDD162 pKa = 3.4HH163 pKa = 6.54NGWSFNDD170 pKa = 3.13QLYY173 pKa = 11.32SMRR176 pKa = 11.84SDD178 pKa = 3.9EE179 pKa = 4.23IYY181 pKa = 10.89EE182 pKa = 4.1GFIYY186 pKa = 9.78MDD188 pKa = 4.9NGDD191 pKa = 3.19NWIGFKK197 pKa = 9.91MSYY200 pKa = 9.54EE201 pKa = 4.09PDD203 pKa = 3.48WIEE206 pKa = 4.58DD207 pKa = 3.86LVIGDD212 pKa = 4.85PDD214 pKa = 3.85DD215 pKa = 4.82SGTSGTLQVGNWGGSNIYY233 pKa = 8.85ATEE236 pKa = 4.26GPGYY240 pKa = 10.43YY241 pKa = 9.73RR242 pKa = 11.84IRR244 pKa = 11.84ANLVNNTISILKK256 pKa = 8.09TEE258 pKa = 4.07WAITGDD264 pKa = 3.61FNGWSFTPLVYY275 pKa = 9.83DD276 pKa = 3.69TDD278 pKa = 4.17DD279 pKa = 4.12KK280 pKa = 11.57VWSATIDD287 pKa = 3.34MTAGGFKK294 pKa = 10.56FIANEE299 pKa = 3.73DD300 pKa = 3.22WSLVFGDD307 pKa = 4.69DD308 pKa = 4.61DD309 pKa = 5.51LNGVLDD315 pKa = 4.95PGRR318 pKa = 11.84DD319 pKa = 3.64DD320 pKa = 4.75NNIQITEE327 pKa = 4.32DD328 pKa = 3.32GNYY331 pKa = 9.28TIIMDD336 pKa = 4.49LSGAPYY342 pKa = 10.12VYY344 pKa = 10.53SIVKK348 pKa = 9.73NN349 pKa = 3.77
Molecular weight: 38.68 kDa
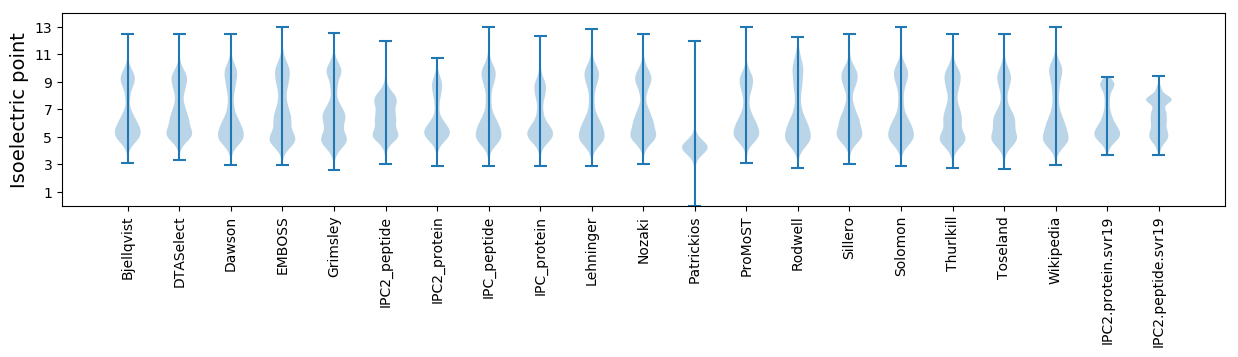
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T5HHU8|A0A1T5HHU8_9BACT Uncharacterized protein OS=Alkalitalea saponilacus OX=889453 GN=SAMN03080601_02328 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.98HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MATANGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.77KK41 pKa = 10.08LTVSDD46 pKa = 3.89EE47 pKa = 4.47RR48 pKa = 11.84KK49 pKa = 10.13HH50 pKa = 5.97KK51 pKa = 10.83AA52 pKa = 3.04
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.98HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MATANGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.77KK41 pKa = 10.08LTVSDD46 pKa = 3.89EE47 pKa = 4.47RR48 pKa = 11.84KK49 pKa = 10.13HH50 pKa = 5.97KK51 pKa = 10.83AA52 pKa = 3.04
Molecular weight: 6.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322236 |
39 |
6369 |
371.0 |
41.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.208 ± 0.04 | 0.786 ± 0.016 |
5.514 ± 0.033 | 6.775 ± 0.039 |
5.218 ± 0.029 | 6.853 ± 0.052 |
2.098 ± 0.017 | 7.733 ± 0.035 |
5.824 ± 0.063 | 9.128 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.52 ± 0.021 | 5.796 ± 0.037 |
3.843 ± 0.024 | 3.356 ± 0.021 |
4.709 ± 0.03 | 6.68 ± 0.031 |
5.263 ± 0.04 | 6.495 ± 0.037 |
1.259 ± 0.014 | 3.944 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
