
Hyella patelloides LEGE 07179
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Pleurocapsales; Hyellaceae; Hyella; Hyella patelloides
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
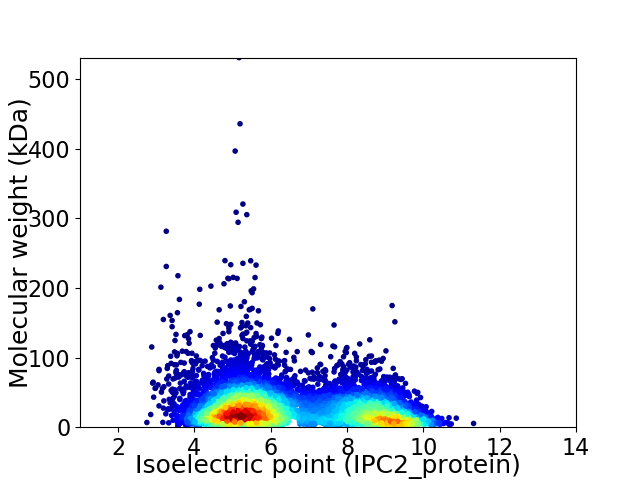
Virtual 2D-PAGE plot for 7698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A563VS95|A0A563VS95_9CYAN Sugar transferase OS=Hyella patelloides LEGE 07179 OX=945734 GN=H1P_2540001 PE=3 SV=1
MM1 pKa = 8.0FIGLCCNQKK10 pKa = 10.38IIINLSVRR18 pKa = 11.84IMLNRR23 pKa = 11.84VFNSLSFKK31 pKa = 10.85AIALSSFFVICHH43 pKa = 5.09VGEE46 pKa = 4.54AKK48 pKa = 10.39AQNTLEE54 pKa = 4.29VPSTDD59 pKa = 3.76YY60 pKa = 10.73QDD62 pKa = 4.94EE63 pKa = 4.4MGQVTNVNQLRR74 pKa = 11.84DD75 pKa = 3.68VSPADD80 pKa = 3.24WSYY83 pKa = 10.69EE84 pKa = 3.83ALRR87 pKa = 11.84SLVDD91 pKa = 3.76RR92 pKa = 11.84YY93 pKa = 11.19GCIAGFPNQAYY104 pKa = 9.82RR105 pKa = 11.84GSQALSRR112 pKa = 11.84YY113 pKa = 8.62EE114 pKa = 3.76FAAGLNSCLNQIEE127 pKa = 4.27RR128 pKa = 11.84LIASSEE134 pKa = 4.0AVVRR138 pKa = 11.84EE139 pKa = 4.69DD140 pKa = 3.03IDD142 pKa = 3.92TINRR146 pKa = 11.84LTQEE150 pKa = 4.41FEE152 pKa = 4.45SEE154 pKa = 3.93LATIGGRR161 pKa = 11.84IDD163 pKa = 3.52SLEE166 pKa = 3.91SRR168 pKa = 11.84TAFLEE173 pKa = 4.26DD174 pKa = 3.47NQFSTTTKK182 pKa = 10.6LNAQVVSYY190 pKa = 11.24VLGTFGDD197 pKa = 4.03EE198 pKa = 4.46RR199 pKa = 11.84PDD201 pKa = 3.49GTDD204 pKa = 2.76VDD206 pKa = 4.25DD207 pKa = 5.35QITFSSRR214 pKa = 11.84VRR216 pKa = 11.84LNFDD220 pKa = 3.23SSFTGEE226 pKa = 3.77DD227 pKa = 3.55FLRR230 pKa = 11.84VRR232 pKa = 11.84IQARR236 pKa = 11.84NITSPAEE243 pKa = 4.08SGGTNSLALNMEE255 pKa = 4.44GDD257 pKa = 3.8SEE259 pKa = 4.99NNVEE263 pKa = 5.91LDD265 pKa = 3.18DD266 pKa = 4.83LYY268 pKa = 11.52YY269 pKa = 10.16IFPVTEE275 pKa = 3.87QVTATVGTNGVDD287 pKa = 2.95VDD289 pKa = 4.72EE290 pKa = 5.49IFDD293 pKa = 3.76VVPTMGIAYY302 pKa = 8.05DD303 pKa = 3.77ALSLYY308 pKa = 10.37SAYY311 pKa = 11.26NNLIYY316 pKa = 11.03SNGNGGGAGIGASLEE331 pKa = 4.11FVEE334 pKa = 5.35WADD337 pKa = 3.9LQIGYY342 pKa = 7.93WAPNPADD349 pKa = 3.98PDD351 pKa = 3.63EE352 pKa = 4.91GNGLFNGNYY361 pKa = 7.73TAGANLNLSFLDD373 pKa = 3.28EE374 pKa = 4.39RR375 pKa = 11.84LNVAVAYY382 pKa = 10.53LRR384 pKa = 11.84GYY386 pKa = 9.21QGQGSGYY393 pKa = 10.23DD394 pKa = 3.18LGGFTGTDD402 pKa = 3.45GATDD406 pKa = 3.97PFDD409 pKa = 4.55DD410 pKa = 5.03QANSSNNFGVATSFQVFEE428 pKa = 4.01QLTVGGYY435 pKa = 9.89FGYY438 pKa = 9.05ATASLVGDD446 pKa = 4.78DD447 pKa = 4.87DD448 pKa = 4.33TNADD452 pKa = 3.34ILTWNAYY459 pKa = 7.73LTYY462 pKa = 10.46TDD464 pKa = 4.47LLKK467 pKa = 10.94EE468 pKa = 4.15GSTALVSFGQPPSLIDD484 pKa = 3.69SEE486 pKa = 4.56GDD488 pKa = 3.26ALADD492 pKa = 4.86DD493 pKa = 4.8EE494 pKa = 4.88DD495 pKa = 4.66TPFLLNVEE503 pKa = 4.25YY504 pKa = 10.4QYY506 pKa = 11.72QLNDD510 pKa = 4.94FIQLTPGAYY519 pKa = 10.3VLFNPNGNSDD529 pKa = 3.32NDD531 pKa = 4.2TIYY534 pKa = 10.89VGTVRR539 pKa = 11.84TIFSFF544 pKa = 3.57
MM1 pKa = 8.0FIGLCCNQKK10 pKa = 10.38IIINLSVRR18 pKa = 11.84IMLNRR23 pKa = 11.84VFNSLSFKK31 pKa = 10.85AIALSSFFVICHH43 pKa = 5.09VGEE46 pKa = 4.54AKK48 pKa = 10.39AQNTLEE54 pKa = 4.29VPSTDD59 pKa = 3.76YY60 pKa = 10.73QDD62 pKa = 4.94EE63 pKa = 4.4MGQVTNVNQLRR74 pKa = 11.84DD75 pKa = 3.68VSPADD80 pKa = 3.24WSYY83 pKa = 10.69EE84 pKa = 3.83ALRR87 pKa = 11.84SLVDD91 pKa = 3.76RR92 pKa = 11.84YY93 pKa = 11.19GCIAGFPNQAYY104 pKa = 9.82RR105 pKa = 11.84GSQALSRR112 pKa = 11.84YY113 pKa = 8.62EE114 pKa = 3.76FAAGLNSCLNQIEE127 pKa = 4.27RR128 pKa = 11.84LIASSEE134 pKa = 4.0AVVRR138 pKa = 11.84EE139 pKa = 4.69DD140 pKa = 3.03IDD142 pKa = 3.92TINRR146 pKa = 11.84LTQEE150 pKa = 4.41FEE152 pKa = 4.45SEE154 pKa = 3.93LATIGGRR161 pKa = 11.84IDD163 pKa = 3.52SLEE166 pKa = 3.91SRR168 pKa = 11.84TAFLEE173 pKa = 4.26DD174 pKa = 3.47NQFSTTTKK182 pKa = 10.6LNAQVVSYY190 pKa = 11.24VLGTFGDD197 pKa = 4.03EE198 pKa = 4.46RR199 pKa = 11.84PDD201 pKa = 3.49GTDD204 pKa = 2.76VDD206 pKa = 4.25DD207 pKa = 5.35QITFSSRR214 pKa = 11.84VRR216 pKa = 11.84LNFDD220 pKa = 3.23SSFTGEE226 pKa = 3.77DD227 pKa = 3.55FLRR230 pKa = 11.84VRR232 pKa = 11.84IQARR236 pKa = 11.84NITSPAEE243 pKa = 4.08SGGTNSLALNMEE255 pKa = 4.44GDD257 pKa = 3.8SEE259 pKa = 4.99NNVEE263 pKa = 5.91LDD265 pKa = 3.18DD266 pKa = 4.83LYY268 pKa = 11.52YY269 pKa = 10.16IFPVTEE275 pKa = 3.87QVTATVGTNGVDD287 pKa = 2.95VDD289 pKa = 4.72EE290 pKa = 5.49IFDD293 pKa = 3.76VVPTMGIAYY302 pKa = 8.05DD303 pKa = 3.77ALSLYY308 pKa = 10.37SAYY311 pKa = 11.26NNLIYY316 pKa = 11.03SNGNGGGAGIGASLEE331 pKa = 4.11FVEE334 pKa = 5.35WADD337 pKa = 3.9LQIGYY342 pKa = 7.93WAPNPADD349 pKa = 3.98PDD351 pKa = 3.63EE352 pKa = 4.91GNGLFNGNYY361 pKa = 7.73TAGANLNLSFLDD373 pKa = 3.28EE374 pKa = 4.39RR375 pKa = 11.84LNVAVAYY382 pKa = 10.53LRR384 pKa = 11.84GYY386 pKa = 9.21QGQGSGYY393 pKa = 10.23DD394 pKa = 3.18LGGFTGTDD402 pKa = 3.45GATDD406 pKa = 3.97PFDD409 pKa = 4.55DD410 pKa = 5.03QANSSNNFGVATSFQVFEE428 pKa = 4.01QLTVGGYY435 pKa = 9.89FGYY438 pKa = 9.05ATASLVGDD446 pKa = 4.78DD447 pKa = 4.87DD448 pKa = 4.33TNADD452 pKa = 3.34ILTWNAYY459 pKa = 7.73LTYY462 pKa = 10.46TDD464 pKa = 4.47LLKK467 pKa = 10.94EE468 pKa = 4.15GSTALVSFGQPPSLIDD484 pKa = 3.69SEE486 pKa = 4.56GDD488 pKa = 3.26ALADD492 pKa = 4.86DD493 pKa = 4.8EE494 pKa = 4.88DD495 pKa = 4.66TPFLLNVEE503 pKa = 4.25YY504 pKa = 10.4QYY506 pKa = 11.72QLNDD510 pKa = 4.94FIQLTPGAYY519 pKa = 10.3VLFNPNGNSDD529 pKa = 3.32NDD531 pKa = 4.2TIYY534 pKa = 10.89VGTVRR539 pKa = 11.84TIFSFF544 pKa = 3.57
Molecular weight: 59.3 kDa
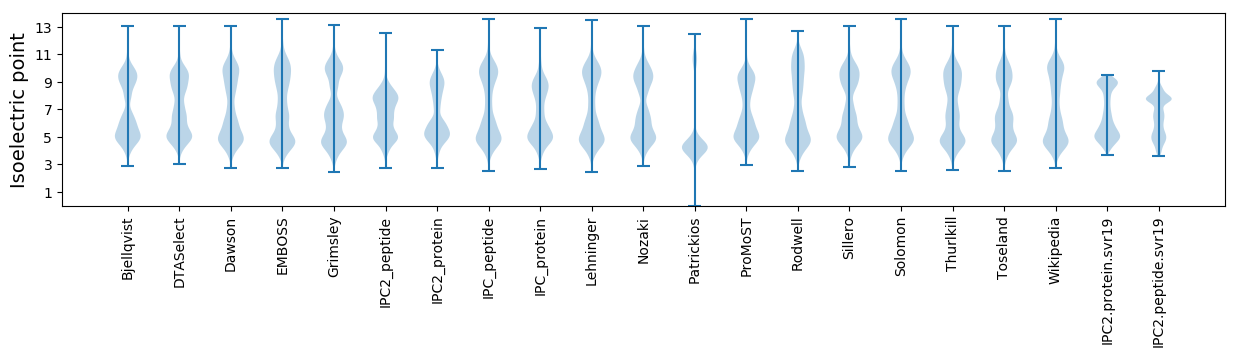
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A563VKW9|A0A563VKW9_9CYAN Toxin of the ChpA-ChpR toxin-antitoxin system endoribonuclease OS=Hyella patelloides LEGE 07179 OX=945734 GN=chpA PE=3 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.13RR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.52TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.21KK38 pKa = 9.62GRR40 pKa = 11.84HH41 pKa = 4.8RR42 pKa = 11.84LAVV45 pKa = 3.37
MM1 pKa = 7.55TKK3 pKa = 9.13RR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.52TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.21KK38 pKa = 9.62GRR40 pKa = 11.84HH41 pKa = 4.8RR42 pKa = 11.84LAVV45 pKa = 3.37
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2095954 |
20 |
4713 |
272.3 |
30.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.294 ± 0.026 | 1.015 ± 0.01 |
5.135 ± 0.032 | 6.54 ± 0.033 |
4.126 ± 0.023 | 6.235 ± 0.039 |
1.712 ± 0.015 | 7.543 ± 0.024 |
5.607 ± 0.037 | 10.782 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.778 ± 0.011 | 5.083 ± 0.028 |
4.095 ± 0.021 | 5.124 ± 0.032 |
4.663 ± 0.023 | 6.827 ± 0.023 |
5.751 ± 0.028 | 6.079 ± 0.02 |
1.366 ± 0.013 | 3.245 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
