
Jiangella rhizosphaerae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Jiangella
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
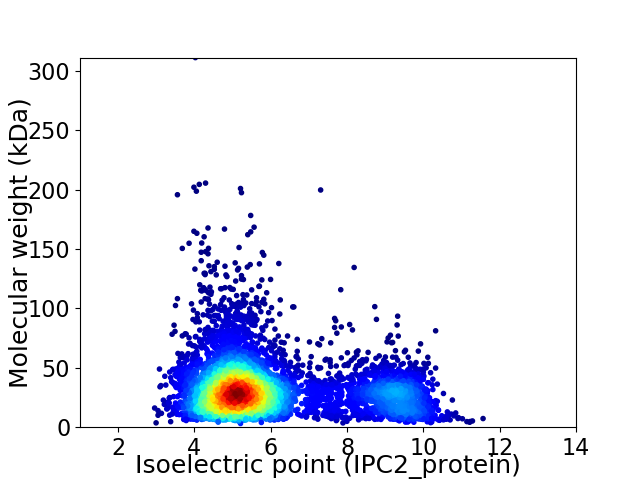
Virtual 2D-PAGE plot for 6027 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418KPU2|A0A418KPU2_9ACTN Iron ABC transporter permease OS=Jiangella rhizosphaerae OX=2293569 GN=DY240_14775 PE=3 SV=1
MM1 pKa = 7.43PVSPRR6 pKa = 11.84RR7 pKa = 11.84SVVLGTVAIALALGACGGGDD27 pKa = 4.18DD28 pKa = 4.32GGEE31 pKa = 3.88AAGEE35 pKa = 4.1GDD37 pKa = 3.29NVVTVRR43 pKa = 11.84GCNPQSAFVPANSNEE58 pKa = 4.1VCSGDD63 pKa = 3.73ILDD66 pKa = 3.97QVFSFLIRR74 pKa = 11.84YY75 pKa = 8.34DD76 pKa = 3.7AEE78 pKa = 4.22TGEE81 pKa = 4.33PTNEE85 pKa = 3.36IAADD89 pKa = 3.12ITTPDD94 pKa = 4.02NGLTWTITLEE104 pKa = 4.66DD105 pKa = 3.48GWTFHH110 pKa = 7.46DD111 pKa = 5.33GSPVTAQSFVDD122 pKa = 3.58AWNWAAYY129 pKa = 9.73GDD131 pKa = 3.99NATLNSYY138 pKa = 10.16FFEE141 pKa = 5.46PIQGYY146 pKa = 10.57AEE148 pKa = 4.1VQNQVDD154 pKa = 3.59ADD156 pKa = 3.96GNPIEE161 pKa = 4.71GTAQAEE167 pKa = 4.51TMSGLEE173 pKa = 4.09VVDD176 pKa = 5.14DD177 pKa = 3.7LTFNVTLTQAEE188 pKa = 4.59SQFPMRR194 pKa = 11.84LGYY197 pKa = 8.26TAFAPLPEE205 pKa = 4.56SFYY208 pKa = 10.99EE209 pKa = 4.22DD210 pKa = 3.64PEE212 pKa = 4.91AFGQHH217 pKa = 7.0PIGTGPFQFEE227 pKa = 3.93SWEE230 pKa = 4.01PNIDD234 pKa = 3.49VRR236 pKa = 11.84LTAYY240 pKa = 9.64PDD242 pKa = 3.42YY243 pKa = 11.2NGDD246 pKa = 3.61VKK248 pKa = 10.98PSIDD252 pKa = 3.16GAIYY256 pKa = 10.2RR257 pKa = 11.84IYY259 pKa = 10.84EE260 pKa = 4.2NDD262 pKa = 3.31DD263 pKa = 2.99AAYY266 pKa = 10.95NDD268 pKa = 4.05LQADD272 pKa = 3.79QLDD275 pKa = 4.16IMPLLPTSALAGDD288 pKa = 4.72TYY290 pKa = 10.94RR291 pKa = 11.84IDD293 pKa = 3.51LGDD296 pKa = 3.47RR297 pKa = 11.84FISRR301 pKa = 11.84EE302 pKa = 3.82TGVFEE307 pKa = 5.15TITFAPASVDD317 pKa = 3.23EE318 pKa = 4.8RR319 pKa = 11.84FADD322 pKa = 3.55PRR324 pKa = 11.84IRR326 pKa = 11.84QAISMAINRR335 pKa = 11.84EE336 pKa = 4.09EE337 pKa = 3.75IVEE340 pKa = 4.21QIFSGARR347 pKa = 11.84TPATGWVSPVVVNGYY362 pKa = 9.52VEE364 pKa = 4.66GACGEE369 pKa = 4.29FCTYY373 pKa = 11.12DD374 pKa = 3.24PGAAQDD380 pKa = 5.34LYY382 pKa = 11.21AQTDD386 pKa = 4.79GIDD389 pKa = 3.6GPFPLSYY396 pKa = 10.81NADD399 pKa = 3.69SDD401 pKa = 3.69HH402 pKa = 6.55KK403 pKa = 10.19AWVEE407 pKa = 3.94AVCNQIDD414 pKa = 3.44ATLPVDD420 pKa = 4.17CVPTPFVDD428 pKa = 4.01LATFRR433 pKa = 11.84NEE435 pKa = 3.17ITNRR439 pKa = 11.84EE440 pKa = 4.11LTGMFRR446 pKa = 11.84TGWQMDD452 pKa = 4.0YY453 pKa = 11.08PSLEE457 pKa = 4.05NFLVPIFATGASANDD472 pKa = 3.52GDD474 pKa = 4.16YY475 pKa = 11.66SNAEE479 pKa = 3.67FDD481 pKa = 3.93QLVEE485 pKa = 3.99EE486 pKa = 4.56AARR489 pKa = 11.84TEE491 pKa = 4.37GEE493 pKa = 4.26AGAEE497 pKa = 4.27LYY499 pKa = 10.47RR500 pKa = 11.84QAEE503 pKa = 3.97ALLRR507 pKa = 11.84NDD509 pKa = 3.73MPAIPLWHH517 pKa = 6.76RR518 pKa = 11.84VNIAGYY524 pKa = 8.69SSHH527 pKa = 7.17VEE529 pKa = 3.94NVQLTVFQTVDD540 pKa = 3.73LLSVTTTGG548 pKa = 3.78
MM1 pKa = 7.43PVSPRR6 pKa = 11.84RR7 pKa = 11.84SVVLGTVAIALALGACGGGDD27 pKa = 4.18DD28 pKa = 4.32GGEE31 pKa = 3.88AAGEE35 pKa = 4.1GDD37 pKa = 3.29NVVTVRR43 pKa = 11.84GCNPQSAFVPANSNEE58 pKa = 4.1VCSGDD63 pKa = 3.73ILDD66 pKa = 3.97QVFSFLIRR74 pKa = 11.84YY75 pKa = 8.34DD76 pKa = 3.7AEE78 pKa = 4.22TGEE81 pKa = 4.33PTNEE85 pKa = 3.36IAADD89 pKa = 3.12ITTPDD94 pKa = 4.02NGLTWTITLEE104 pKa = 4.66DD105 pKa = 3.48GWTFHH110 pKa = 7.46DD111 pKa = 5.33GSPVTAQSFVDD122 pKa = 3.58AWNWAAYY129 pKa = 9.73GDD131 pKa = 3.99NATLNSYY138 pKa = 10.16FFEE141 pKa = 5.46PIQGYY146 pKa = 10.57AEE148 pKa = 4.1VQNQVDD154 pKa = 3.59ADD156 pKa = 3.96GNPIEE161 pKa = 4.71GTAQAEE167 pKa = 4.51TMSGLEE173 pKa = 4.09VVDD176 pKa = 5.14DD177 pKa = 3.7LTFNVTLTQAEE188 pKa = 4.59SQFPMRR194 pKa = 11.84LGYY197 pKa = 8.26TAFAPLPEE205 pKa = 4.56SFYY208 pKa = 10.99EE209 pKa = 4.22DD210 pKa = 3.64PEE212 pKa = 4.91AFGQHH217 pKa = 7.0PIGTGPFQFEE227 pKa = 3.93SWEE230 pKa = 4.01PNIDD234 pKa = 3.49VRR236 pKa = 11.84LTAYY240 pKa = 9.64PDD242 pKa = 3.42YY243 pKa = 11.2NGDD246 pKa = 3.61VKK248 pKa = 10.98PSIDD252 pKa = 3.16GAIYY256 pKa = 10.2RR257 pKa = 11.84IYY259 pKa = 10.84EE260 pKa = 4.2NDD262 pKa = 3.31DD263 pKa = 2.99AAYY266 pKa = 10.95NDD268 pKa = 4.05LQADD272 pKa = 3.79QLDD275 pKa = 4.16IMPLLPTSALAGDD288 pKa = 4.72TYY290 pKa = 10.94RR291 pKa = 11.84IDD293 pKa = 3.51LGDD296 pKa = 3.47RR297 pKa = 11.84FISRR301 pKa = 11.84EE302 pKa = 3.82TGVFEE307 pKa = 5.15TITFAPASVDD317 pKa = 3.23EE318 pKa = 4.8RR319 pKa = 11.84FADD322 pKa = 3.55PRR324 pKa = 11.84IRR326 pKa = 11.84QAISMAINRR335 pKa = 11.84EE336 pKa = 4.09EE337 pKa = 3.75IVEE340 pKa = 4.21QIFSGARR347 pKa = 11.84TPATGWVSPVVVNGYY362 pKa = 9.52VEE364 pKa = 4.66GACGEE369 pKa = 4.29FCTYY373 pKa = 11.12DD374 pKa = 3.24PGAAQDD380 pKa = 5.34LYY382 pKa = 11.21AQTDD386 pKa = 4.79GIDD389 pKa = 3.6GPFPLSYY396 pKa = 10.81NADD399 pKa = 3.69SDD401 pKa = 3.69HH402 pKa = 6.55KK403 pKa = 10.19AWVEE407 pKa = 3.94AVCNQIDD414 pKa = 3.44ATLPVDD420 pKa = 4.17CVPTPFVDD428 pKa = 4.01LATFRR433 pKa = 11.84NEE435 pKa = 3.17ITNRR439 pKa = 11.84EE440 pKa = 4.11LTGMFRR446 pKa = 11.84TGWQMDD452 pKa = 4.0YY453 pKa = 11.08PSLEE457 pKa = 4.05NFLVPIFATGASANDD472 pKa = 3.52GDD474 pKa = 4.16YY475 pKa = 11.66SNAEE479 pKa = 3.67FDD481 pKa = 3.93QLVEE485 pKa = 3.99EE486 pKa = 4.56AARR489 pKa = 11.84TEE491 pKa = 4.37GEE493 pKa = 4.26AGAEE497 pKa = 4.27LYY499 pKa = 10.47RR500 pKa = 11.84QAEE503 pKa = 3.97ALLRR507 pKa = 11.84NDD509 pKa = 3.73MPAIPLWHH517 pKa = 6.76RR518 pKa = 11.84VNIAGYY524 pKa = 8.69SSHH527 pKa = 7.17VEE529 pKa = 3.94NVQLTVFQTVDD540 pKa = 3.73LLSVTTTGG548 pKa = 3.78
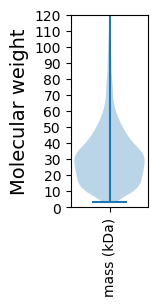
Molecular weight: 59.54 kDa
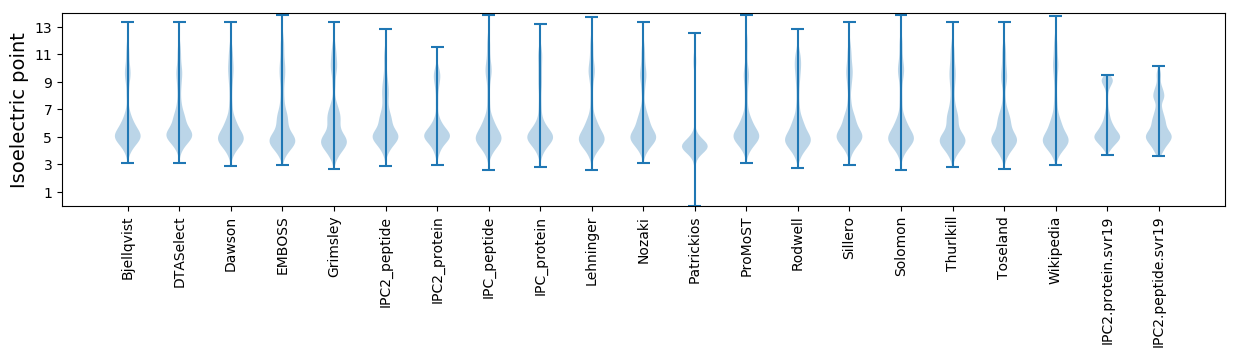
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418KPX7|A0A418KPX7_9ACTN Type II toxin-antitoxin system Phd/YefM family antitoxin OS=Jiangella rhizosphaerae OX=2293569 GN=DY240_14385 PE=3 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.78GRR40 pKa = 11.84AKK42 pKa = 10.73LSAA45 pKa = 3.92
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.78GRR40 pKa = 11.84AKK42 pKa = 10.73LSAA45 pKa = 3.92
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1902621 |
29 |
3103 |
315.7 |
33.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.932 ± 0.049 | 0.622 ± 0.008 |
6.685 ± 0.027 | 5.61 ± 0.033 |
2.787 ± 0.019 | 9.42 ± 0.03 |
2.103 ± 0.015 | 3.313 ± 0.023 |
1.363 ± 0.018 | 10.23 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.013 | 1.686 ± 0.017 |
5.997 ± 0.027 | 2.547 ± 0.015 |
8.017 ± 0.038 | 4.847 ± 0.022 |
6.027 ± 0.028 | 9.477 ± 0.03 |
1.603 ± 0.016 | 2.087 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
