
Volvox carteri f. nagariensis
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Chlorophyceae; CS clade; Chlamydomonadales; Volvocaceae; Volvox; Volvox carteri
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
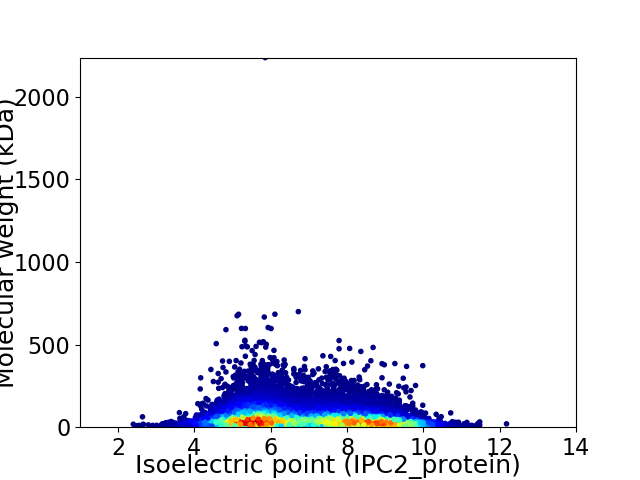
Virtual 2D-PAGE plot for 14335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8TXQ2|D8TXQ2_VOLCA Uncharacterized protein OS=Volvox carteri f. nagariensis OX=3068 GN=VOLCADRAFT_105002 PE=4 SV=1
MM1 pKa = 7.53GKK3 pKa = 9.82YY4 pKa = 9.21AQLVIGPAGCGKK16 pKa = 8.65STYY19 pKa = 10.06CNHH22 pKa = 7.24LYY24 pKa = 9.39EE25 pKa = 4.86HH26 pKa = 6.75CQAIKK31 pKa = 10.53RR32 pKa = 11.84SVHH35 pKa = 5.37VVNLDD40 pKa = 3.43PAAEE44 pKa = 4.1AFQYY48 pKa = 10.15PVSLDD53 pKa = 3.26IRR55 pKa = 11.84DD56 pKa = 4.07LVCLEE61 pKa = 4.6DD62 pKa = 3.82VMEE65 pKa = 4.44EE66 pKa = 4.52LGLGPNGGLLYY77 pKa = 10.84CMEE80 pKa = 4.19YY81 pKa = 11.05LEE83 pKa = 5.95DD84 pKa = 5.57NLHH87 pKa = 5.78EE88 pKa = 4.25WLGEE92 pKa = 3.93EE93 pKa = 4.37LEE95 pKa = 4.78GYY97 pKa = 10.47GDD99 pKa = 3.54EE100 pKa = 5.19DD101 pKa = 4.61YY102 pKa = 11.56LVFDD106 pKa = 4.74CPGQIEE112 pKa = 5.27LYY114 pKa = 9.65NHH116 pKa = 6.79LSVFRR121 pKa = 11.84SFVDD125 pKa = 3.84FLKK128 pKa = 10.89NDD130 pKa = 3.12GWSVCVVYY138 pKa = 10.83CLDD141 pKa = 3.18AHH143 pKa = 6.55FVTDD147 pKa = 3.28VAKK150 pKa = 10.73FMAGALQALAAMVKK164 pKa = 10.63LEE166 pKa = 4.29LPHH169 pKa = 8.09VNILTKK175 pKa = 10.44VDD177 pKa = 3.4LMEE180 pKa = 5.67DD181 pKa = 3.63KK182 pKa = 11.25NHH184 pKa = 6.76LDD186 pKa = 3.33EE187 pKa = 6.13FLFPDD192 pKa = 4.48PEE194 pKa = 4.76LLLHH198 pKa = 6.19QLAASTGPRR207 pKa = 11.84FRR209 pKa = 11.84QLNRR213 pKa = 11.84AMGGLLEE220 pKa = 4.39EE221 pKa = 4.77FSLVSFLPLDD231 pKa = 3.51ITDD234 pKa = 4.13EE235 pKa = 4.28DD236 pKa = 4.62SIADD240 pKa = 3.44ILGQIDD246 pKa = 3.42MAIQYY251 pKa = 10.78GEE253 pKa = 4.07DD254 pKa = 3.36AEE256 pKa = 4.18PRR258 pKa = 11.84IRR260 pKa = 11.84DD261 pKa = 3.65EE262 pKa = 5.56LEE264 pKa = 4.61DD265 pKa = 5.27GGDD268 pKa = 4.71DD269 pKa = 5.47DD270 pKa = 6.97DD271 pKa = 7.51DD272 pKa = 6.64DD273 pKa = 6.4GGGDD277 pKa = 3.7GGDD280 pKa = 4.53DD281 pKa = 3.09II282 pKa = 6.89
MM1 pKa = 7.53GKK3 pKa = 9.82YY4 pKa = 9.21AQLVIGPAGCGKK16 pKa = 8.65STYY19 pKa = 10.06CNHH22 pKa = 7.24LYY24 pKa = 9.39EE25 pKa = 4.86HH26 pKa = 6.75CQAIKK31 pKa = 10.53RR32 pKa = 11.84SVHH35 pKa = 5.37VVNLDD40 pKa = 3.43PAAEE44 pKa = 4.1AFQYY48 pKa = 10.15PVSLDD53 pKa = 3.26IRR55 pKa = 11.84DD56 pKa = 4.07LVCLEE61 pKa = 4.6DD62 pKa = 3.82VMEE65 pKa = 4.44EE66 pKa = 4.52LGLGPNGGLLYY77 pKa = 10.84CMEE80 pKa = 4.19YY81 pKa = 11.05LEE83 pKa = 5.95DD84 pKa = 5.57NLHH87 pKa = 5.78EE88 pKa = 4.25WLGEE92 pKa = 3.93EE93 pKa = 4.37LEE95 pKa = 4.78GYY97 pKa = 10.47GDD99 pKa = 3.54EE100 pKa = 5.19DD101 pKa = 4.61YY102 pKa = 11.56LVFDD106 pKa = 4.74CPGQIEE112 pKa = 5.27LYY114 pKa = 9.65NHH116 pKa = 6.79LSVFRR121 pKa = 11.84SFVDD125 pKa = 3.84FLKK128 pKa = 10.89NDD130 pKa = 3.12GWSVCVVYY138 pKa = 10.83CLDD141 pKa = 3.18AHH143 pKa = 6.55FVTDD147 pKa = 3.28VAKK150 pKa = 10.73FMAGALQALAAMVKK164 pKa = 10.63LEE166 pKa = 4.29LPHH169 pKa = 8.09VNILTKK175 pKa = 10.44VDD177 pKa = 3.4LMEE180 pKa = 5.67DD181 pKa = 3.63KK182 pKa = 11.25NHH184 pKa = 6.76LDD186 pKa = 3.33EE187 pKa = 6.13FLFPDD192 pKa = 4.48PEE194 pKa = 4.76LLLHH198 pKa = 6.19QLAASTGPRR207 pKa = 11.84FRR209 pKa = 11.84QLNRR213 pKa = 11.84AMGGLLEE220 pKa = 4.39EE221 pKa = 4.77FSLVSFLPLDD231 pKa = 3.51ITDD234 pKa = 4.13EE235 pKa = 4.28DD236 pKa = 4.62SIADD240 pKa = 3.44ILGQIDD246 pKa = 3.42MAIQYY251 pKa = 10.78GEE253 pKa = 4.07DD254 pKa = 3.36AEE256 pKa = 4.18PRR258 pKa = 11.84IRR260 pKa = 11.84DD261 pKa = 3.65EE262 pKa = 5.56LEE264 pKa = 4.61DD265 pKa = 5.27GGDD268 pKa = 4.71DD269 pKa = 5.47DD270 pKa = 6.97DD271 pKa = 7.51DD272 pKa = 6.64DD273 pKa = 6.4GGGDD277 pKa = 3.7GGDD280 pKa = 4.53DD281 pKa = 3.09II282 pKa = 6.89
Molecular weight: 31.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8TS10|D8TS10_VOLCA Uncharacterized protein (Fragment) OS=Volvox carteri f. nagariensis OX=3068 GN=VOLCADRAFT_59125 PE=4 SV=1
PP1 pKa = 6.77HH2 pKa = 6.72TRR4 pKa = 11.84GPHH7 pKa = 4.23TRR9 pKa = 11.84GPHH12 pKa = 4.27TRR14 pKa = 11.84GPHH17 pKa = 4.27TRR19 pKa = 11.84GPHH22 pKa = 4.27TRR24 pKa = 11.84GPHH27 pKa = 4.27TRR29 pKa = 11.84GPHH32 pKa = 4.21TRR34 pKa = 11.84GAHH37 pKa = 4.37TRR39 pKa = 11.84GPHH42 pKa = 4.48TRR44 pKa = 11.84GAHH47 pKa = 4.37TRR49 pKa = 11.84GPHH52 pKa = 4.48TRR54 pKa = 11.84GPHH57 pKa = 4.27TRR59 pKa = 11.84GPHH62 pKa = 4.27TRR64 pKa = 11.84GPHH67 pKa = 4.27TRR69 pKa = 11.84GPHH72 pKa = 4.49TRR74 pKa = 11.84GPHH77 pKa = 4.71
PP1 pKa = 6.77HH2 pKa = 6.72TRR4 pKa = 11.84GPHH7 pKa = 4.23TRR9 pKa = 11.84GPHH12 pKa = 4.27TRR14 pKa = 11.84GPHH17 pKa = 4.27TRR19 pKa = 11.84GPHH22 pKa = 4.27TRR24 pKa = 11.84GPHH27 pKa = 4.27TRR29 pKa = 11.84GPHH32 pKa = 4.21TRR34 pKa = 11.84GAHH37 pKa = 4.37TRR39 pKa = 11.84GPHH42 pKa = 4.48TRR44 pKa = 11.84GAHH47 pKa = 4.37TRR49 pKa = 11.84GPHH52 pKa = 4.48TRR54 pKa = 11.84GPHH57 pKa = 4.27TRR59 pKa = 11.84GPHH62 pKa = 4.27TRR64 pKa = 11.84GPHH67 pKa = 4.27TRR69 pKa = 11.84GPHH72 pKa = 4.49TRR74 pKa = 11.84GPHH77 pKa = 4.71
Molecular weight: 8.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8189894 |
49 |
22244 |
571.3 |
60.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.297 ± 0.052 | 1.65 ± 0.012 |
4.715 ± 0.013 | 5.211 ± 0.026 |
2.493 ± 0.015 | 9.546 ± 0.035 |
2.26 ± 0.009 | 2.781 ± 0.015 |
3.026 ± 0.019 | 9.227 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.009 | 2.497 ± 0.014 |
7.005 ± 0.032 | 4.462 ± 0.021 |
6.674 ± 0.018 | 7.645 ± 0.025 |
5.307 ± 0.012 | 6.794 ± 0.019 |
1.232 ± 0.007 | 2.103 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
