
Gorilla associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
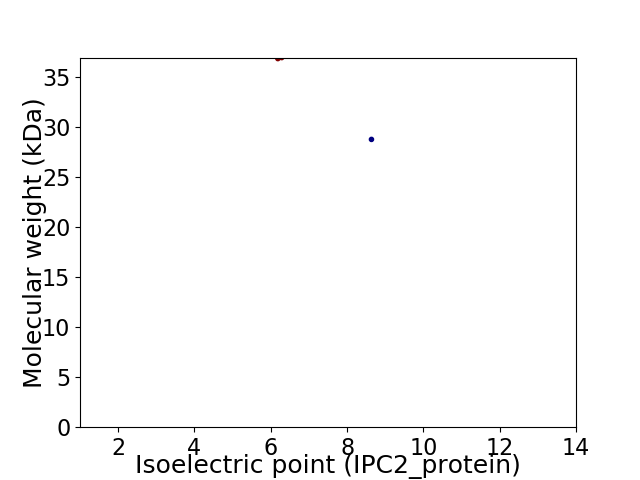
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
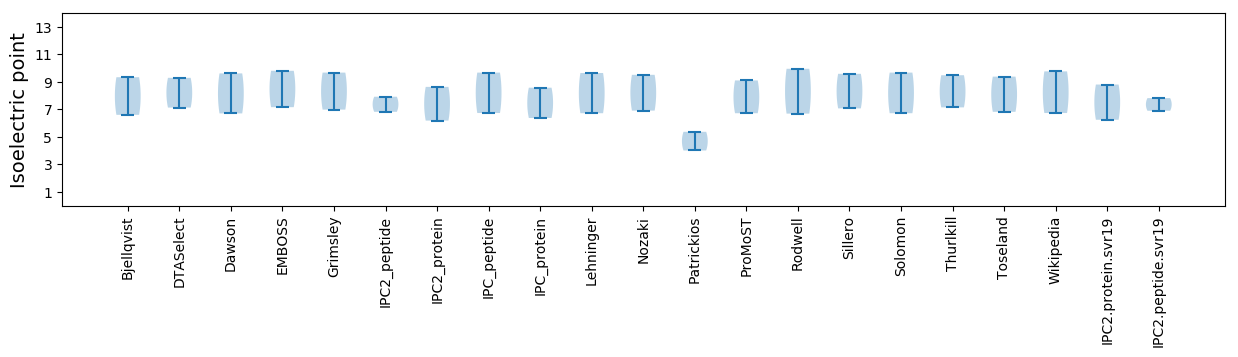
Protein with the lowest isoelectric point:
>tr|A0A0B5GN97|A0A0B5GN97_9VIRU Rep protein OS=Gorilla associated porprismacovirus 1 OX=2170111 GN=rep PE=4 SV=1
MM1 pKa = 6.39MAFRR5 pKa = 11.84KK6 pKa = 9.5RR7 pKa = 11.84HH8 pKa = 4.43GSKK11 pKa = 10.24VLHH14 pKa = 6.69QKK16 pKa = 8.52FQWFFDD22 pKa = 3.94LQTKK26 pKa = 9.58ADD28 pKa = 3.74EE29 pKa = 4.13MQIMEE34 pKa = 4.47VQAGGYY40 pKa = 9.04GVYY43 pKa = 10.16KK44 pKa = 10.34RR45 pKa = 11.84LYY47 pKa = 9.09PFFSAFKK54 pKa = 8.38YY55 pKa = 10.37YY56 pKa = 10.95KK57 pKa = 10.56LGGVKK62 pKa = 9.37MKK64 pKa = 9.33MIPASTLPVDD74 pKa = 3.87PTGLSYY80 pKa = 10.68EE81 pKa = 4.23AGEE84 pKa = 4.26NTVDD88 pKa = 4.85PRR90 pKa = 11.84DD91 pKa = 3.48QLTPGLTRR99 pKa = 11.84ITNGEE104 pKa = 4.09DD105 pKa = 3.47VYY107 pKa = 11.64TDD109 pKa = 3.53LTGLTGDD116 pKa = 3.71QQRR119 pKa = 11.84EE120 pKa = 4.48LYY122 pKa = 10.49EE123 pKa = 4.51SMXIDD128 pKa = 3.37QRR130 pKa = 11.84WFKK133 pKa = 10.18WQLQSGLSRR142 pKa = 11.84YY143 pKa = 8.4ARR145 pKa = 11.84PMYY148 pKa = 8.75WQIGQLHH155 pKa = 6.12QDD157 pKa = 3.76YY158 pKa = 11.0LPGSVRR164 pKa = 11.84NLADD168 pKa = 3.2TSLTDD173 pKa = 3.64DD174 pKa = 4.25CTSITAVYY182 pKa = 10.43DD183 pKa = 3.58GANTSGPSAAPITVAADD200 pKa = 3.18ASDD203 pKa = 3.55PRR205 pKa = 11.84GLFQVGHH212 pKa = 7.01RR213 pKa = 11.84GRR215 pKa = 11.84LGWMPTDD222 pKa = 3.53GVLLKK227 pKa = 9.66GTKK230 pKa = 9.61SGQTGTVHH238 pKa = 6.04AQAVEE243 pKa = 3.92AAIPAVNVFTIVTPPMHH260 pKa = 6.15KK261 pKa = 8.5TNYY264 pKa = 7.64YY265 pKa = 8.29YY266 pKa = 10.56RR267 pKa = 11.84VFVTEE272 pKa = 4.34DD273 pKa = 3.84VYY275 pKa = 11.28FKK277 pKa = 11.2SPVVVGYY284 pKa = 9.58QNYY287 pKa = 9.62RR288 pKa = 11.84SIDD291 pKa = 3.52RR292 pKa = 11.84FVQPQFPVAKK302 pKa = 9.42LPTVNSPATDD312 pKa = 3.44SNTPFPTNDD321 pKa = 3.36GEE323 pKa = 4.31QLPEE327 pKa = 3.82VLII330 pKa = 4.48
MM1 pKa = 6.39MAFRR5 pKa = 11.84KK6 pKa = 9.5RR7 pKa = 11.84HH8 pKa = 4.43GSKK11 pKa = 10.24VLHH14 pKa = 6.69QKK16 pKa = 8.52FQWFFDD22 pKa = 3.94LQTKK26 pKa = 9.58ADD28 pKa = 3.74EE29 pKa = 4.13MQIMEE34 pKa = 4.47VQAGGYY40 pKa = 9.04GVYY43 pKa = 10.16KK44 pKa = 10.34RR45 pKa = 11.84LYY47 pKa = 9.09PFFSAFKK54 pKa = 8.38YY55 pKa = 10.37YY56 pKa = 10.95KK57 pKa = 10.56LGGVKK62 pKa = 9.37MKK64 pKa = 9.33MIPASTLPVDD74 pKa = 3.87PTGLSYY80 pKa = 10.68EE81 pKa = 4.23AGEE84 pKa = 4.26NTVDD88 pKa = 4.85PRR90 pKa = 11.84DD91 pKa = 3.48QLTPGLTRR99 pKa = 11.84ITNGEE104 pKa = 4.09DD105 pKa = 3.47VYY107 pKa = 11.64TDD109 pKa = 3.53LTGLTGDD116 pKa = 3.71QQRR119 pKa = 11.84EE120 pKa = 4.48LYY122 pKa = 10.49EE123 pKa = 4.51SMXIDD128 pKa = 3.37QRR130 pKa = 11.84WFKK133 pKa = 10.18WQLQSGLSRR142 pKa = 11.84YY143 pKa = 8.4ARR145 pKa = 11.84PMYY148 pKa = 8.75WQIGQLHH155 pKa = 6.12QDD157 pKa = 3.76YY158 pKa = 11.0LPGSVRR164 pKa = 11.84NLADD168 pKa = 3.2TSLTDD173 pKa = 3.64DD174 pKa = 4.25CTSITAVYY182 pKa = 10.43DD183 pKa = 3.58GANTSGPSAAPITVAADD200 pKa = 3.18ASDD203 pKa = 3.55PRR205 pKa = 11.84GLFQVGHH212 pKa = 7.01RR213 pKa = 11.84GRR215 pKa = 11.84LGWMPTDD222 pKa = 3.53GVLLKK227 pKa = 9.66GTKK230 pKa = 9.61SGQTGTVHH238 pKa = 6.04AQAVEE243 pKa = 3.92AAIPAVNVFTIVTPPMHH260 pKa = 6.15KK261 pKa = 8.5TNYY264 pKa = 7.64YY265 pKa = 8.29YY266 pKa = 10.56RR267 pKa = 11.84VFVTEE272 pKa = 4.34DD273 pKa = 3.84VYY275 pKa = 11.28FKK277 pKa = 11.2SPVVVGYY284 pKa = 9.58QNYY287 pKa = 9.62RR288 pKa = 11.84SIDD291 pKa = 3.52RR292 pKa = 11.84FVQPQFPVAKK302 pKa = 9.42LPTVNSPATDD312 pKa = 3.44SNTPFPTNDD321 pKa = 3.36GEE323 pKa = 4.31QLPEE327 pKa = 3.82VLII330 pKa = 4.48
Molecular weight: 36.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GN97|A0A0B5GN97_9VIRU Rep protein OS=Gorilla associated porprismacovirus 1 OX=2170111 GN=rep PE=4 SV=1
MM1 pKa = 6.62TQTWMLTVPRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.18TEE15 pKa = 4.0KK16 pKa = 9.83EE17 pKa = 3.46WMGIYY22 pKa = 9.84KK23 pKa = 9.73WLRR26 pKa = 11.84DD27 pKa = 3.58NDD29 pKa = 3.68VHH31 pKa = 6.67KK32 pKa = 9.25WTVAMEE38 pKa = 4.38TGNNGYY44 pKa = 8.37DD45 pKa = 2.66HH46 pKa = 6.22WQIRR50 pKa = 11.84FQVGKK55 pKa = 7.81TFKK58 pKa = 10.19QLKK61 pKa = 9.63KK62 pKa = 8.91EE63 pKa = 4.17WGPKK67 pKa = 9.16AHH69 pKa = 6.94IEE71 pKa = 4.12EE72 pKa = 5.48ASDD75 pKa = 2.77TWEE78 pKa = 4.11YY79 pKa = 10.5EE80 pKa = 4.14RR81 pKa = 11.84KK82 pKa = 10.05SGMFFSSDD90 pKa = 3.26DD91 pKa = 3.56TPEE94 pKa = 3.65VRR96 pKa = 11.84KK97 pKa = 10.25CRR99 pKa = 11.84FGRR102 pKa = 11.84LNWRR106 pKa = 11.84QRR108 pKa = 11.84TIVRR112 pKa = 11.84AVQDD116 pKa = 3.64TNDD119 pKa = 3.5RR120 pKa = 11.84EE121 pKa = 4.28IVVWYY126 pKa = 9.85DD127 pKa = 3.12PNGNKK132 pKa = 9.66GKK134 pKa = 10.11SWLLGHH140 pKa = 7.53LYY142 pKa = 8.31EE143 pKa = 5.31TGQAWVIQAQDD154 pKa = 3.46TVKK157 pKa = 11.18GIIQDD162 pKa = 3.87CASEE166 pKa = 4.62FINHH170 pKa = 5.19GWRR173 pKa = 11.84PIVVIDD179 pKa = 5.32IPRR182 pKa = 11.84TWKK185 pKa = 7.87WTSDD189 pKa = 3.27LYY191 pKa = 11.39VAIEE195 pKa = 4.27RR196 pKa = 11.84IKK198 pKa = 11.05DD199 pKa = 3.64GLIKK203 pKa = 10.46DD204 pKa = 3.5PRR206 pKa = 11.84YY207 pKa = 10.1NSKK210 pKa = 8.3TVHH213 pKa = 4.97IRR215 pKa = 11.84GVKK218 pKa = 9.98VLITCNTMPTRR229 pKa = 11.84DD230 pKa = 4.07KK231 pKa = 11.2LSADD235 pKa = 2.71RR236 pKa = 11.84WVIVEE241 pKa = 4.01LL242 pKa = 4.3
MM1 pKa = 6.62TQTWMLTVPRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.18TEE15 pKa = 4.0KK16 pKa = 9.83EE17 pKa = 3.46WMGIYY22 pKa = 9.84KK23 pKa = 9.73WLRR26 pKa = 11.84DD27 pKa = 3.58NDD29 pKa = 3.68VHH31 pKa = 6.67KK32 pKa = 9.25WTVAMEE38 pKa = 4.38TGNNGYY44 pKa = 8.37DD45 pKa = 2.66HH46 pKa = 6.22WQIRR50 pKa = 11.84FQVGKK55 pKa = 7.81TFKK58 pKa = 10.19QLKK61 pKa = 9.63KK62 pKa = 8.91EE63 pKa = 4.17WGPKK67 pKa = 9.16AHH69 pKa = 6.94IEE71 pKa = 4.12EE72 pKa = 5.48ASDD75 pKa = 2.77TWEE78 pKa = 4.11YY79 pKa = 10.5EE80 pKa = 4.14RR81 pKa = 11.84KK82 pKa = 10.05SGMFFSSDD90 pKa = 3.26DD91 pKa = 3.56TPEE94 pKa = 3.65VRR96 pKa = 11.84KK97 pKa = 10.25CRR99 pKa = 11.84FGRR102 pKa = 11.84LNWRR106 pKa = 11.84QRR108 pKa = 11.84TIVRR112 pKa = 11.84AVQDD116 pKa = 3.64TNDD119 pKa = 3.5RR120 pKa = 11.84EE121 pKa = 4.28IVVWYY126 pKa = 9.85DD127 pKa = 3.12PNGNKK132 pKa = 9.66GKK134 pKa = 10.11SWLLGHH140 pKa = 7.53LYY142 pKa = 8.31EE143 pKa = 5.31TGQAWVIQAQDD154 pKa = 3.46TVKK157 pKa = 11.18GIIQDD162 pKa = 3.87CASEE166 pKa = 4.62FINHH170 pKa = 5.19GWRR173 pKa = 11.84PIVVIDD179 pKa = 5.32IPRR182 pKa = 11.84TWKK185 pKa = 7.87WTSDD189 pKa = 3.27LYY191 pKa = 11.39VAIEE195 pKa = 4.27RR196 pKa = 11.84IKK198 pKa = 11.05DD199 pKa = 3.64GLIKK203 pKa = 10.46DD204 pKa = 3.5PRR206 pKa = 11.84YY207 pKa = 10.1NSKK210 pKa = 8.3TVHH213 pKa = 4.97IRR215 pKa = 11.84GVKK218 pKa = 9.98VLITCNTMPTRR229 pKa = 11.84DD230 pKa = 4.07KK231 pKa = 11.2LSADD235 pKa = 2.71RR236 pKa = 11.84WVIVEE241 pKa = 4.01LL242 pKa = 4.3
Molecular weight: 28.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
572 |
242 |
330 |
286.0 |
32.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.42 ± 1.032 | 0.699 ± 0.328 |
6.643 ± 0.232 | 4.371 ± 0.859 |
3.671 ± 0.724 | 7.343 ± 0.695 |
2.098 ± 0.232 | 5.07 ± 1.438 |
6.119 ± 1.303 | 6.294 ± 0.811 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.797 ± 0.193 | 3.497 ± 0.386 |
5.42 ± 1.283 | 5.42 ± 0.782 |
6.119 ± 1.052 | 4.72 ± 0.608 |
8.217 ± 0.222 | 8.042 ± 0.116 |
3.497 ± 1.64 | 4.371 ± 0.897 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
