
Flammulina velutipes browning virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
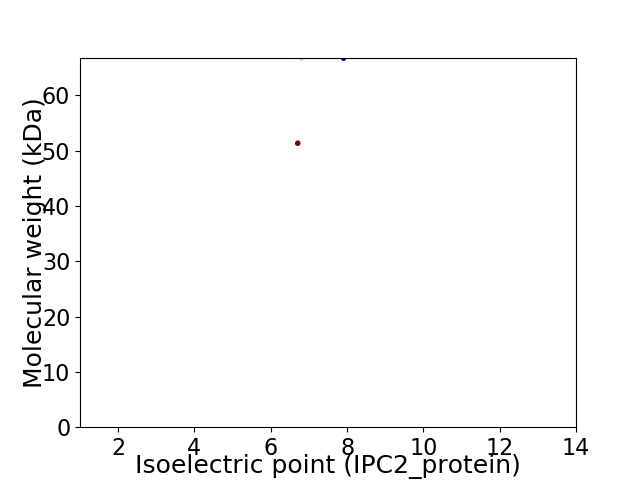
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0STM1|C0STM1_9VIRU Coat protein OS=Flammulina velutipes browning virus OX=568090 GN=CP1 PE=4 SV=1
MM1 pKa = 7.5SPPRR5 pKa = 11.84FPKK8 pKa = 10.6PPVTPSASQTADD20 pKa = 3.31PLGTRR25 pKa = 11.84DD26 pKa = 3.47VDD28 pKa = 4.19SEE30 pKa = 4.41SSKK33 pKa = 10.9PKK35 pKa = 10.22LVNSRR40 pKa = 11.84PQKK43 pKa = 9.6AVPRR47 pKa = 11.84AGPTSAPSVSDD58 pKa = 3.59ANPIYY63 pKa = 10.37HH64 pKa = 6.69AVSNFPYY71 pKa = 10.27YY72 pKa = 9.8PHH74 pKa = 6.53QRR76 pKa = 11.84LDD78 pKa = 2.94TSTFVPCTQVLFAVLAHH95 pKa = 6.78MDD97 pKa = 3.9NIMCTTYY104 pKa = 10.5SWTQSQPAWHH114 pKa = 6.94PCVSRR119 pKa = 11.84LYY121 pKa = 10.59FSTLVYY127 pKa = 9.96YY128 pKa = 10.22QILRR132 pKa = 11.84AMDD135 pKa = 4.09AARR138 pKa = 11.84IITVLQKK145 pKa = 10.76EE146 pKa = 4.26LLNFLEE152 pKa = 4.3HH153 pKa = 6.99TIPASALPIPGPLAPFLRR171 pKa = 11.84SPFLLFNWHH180 pKa = 6.01GWLLQCMSNHH190 pKa = 5.76TXWPHH195 pKa = 5.0HH196 pKa = 5.97QKK198 pKa = 9.99IKK200 pKa = 10.74YY201 pKa = 9.14NISSAFNNLLPNLVHH216 pKa = 6.95AVDD219 pKa = 3.7SAYY222 pKa = 11.3SFATDD227 pKa = 2.86ATYY230 pKa = 11.23VFRR233 pKa = 11.84SAVNDD238 pKa = 3.81LNVKK242 pKa = 9.39MEE244 pKa = 4.21ATDD247 pKa = 3.77PDD249 pKa = 3.69MPAAEE254 pKa = 4.07TRR256 pKa = 11.84HH257 pKa = 5.8WNSAMMDD264 pKa = 3.55PVYY267 pKa = 10.65RR268 pKa = 11.84NDD270 pKa = 3.03MYY272 pKa = 11.1IPTRR276 pKa = 11.84VAQNFAAFSAPLGLPLRR293 pKa = 11.84YY294 pKa = 9.81NYY296 pKa = 10.87GPDD299 pKa = 3.32TDD301 pKa = 4.0IGLSHH306 pKa = 7.55FLYY309 pKa = 10.87LSGGKK314 pKa = 9.0HH315 pKa = 7.0DD316 pKa = 4.29YY317 pKa = 11.07LNRR320 pKa = 11.84LAPVFARR327 pKa = 11.84YY328 pKa = 8.6SQYY331 pKa = 10.66FVNSKK336 pKa = 10.14SLQEE340 pKa = 4.04CSPVGSAPGQINGTLTDD357 pKa = 5.83AIPVPTISGTTVYY370 pKa = 10.22TPSTLAALVTPWSSTTIYY388 pKa = 9.47THH390 pKa = 6.58VNLSLPEE397 pKa = 4.07FFEE400 pKa = 4.38HH401 pKa = 6.5QAALTNVNVTEE412 pKa = 4.73HH413 pKa = 6.32GAAPTFGTTATTLNGSFWRR432 pKa = 11.84MKK434 pKa = 8.42PHH436 pKa = 5.8YY437 pKa = 9.82RR438 pKa = 11.84EE439 pKa = 4.18SPPINVTKK447 pKa = 10.8NLFNIISNEE456 pKa = 3.72MFAEE460 pKa = 4.44PTSS463 pKa = 3.57
MM1 pKa = 7.5SPPRR5 pKa = 11.84FPKK8 pKa = 10.6PPVTPSASQTADD20 pKa = 3.31PLGTRR25 pKa = 11.84DD26 pKa = 3.47VDD28 pKa = 4.19SEE30 pKa = 4.41SSKK33 pKa = 10.9PKK35 pKa = 10.22LVNSRR40 pKa = 11.84PQKK43 pKa = 9.6AVPRR47 pKa = 11.84AGPTSAPSVSDD58 pKa = 3.59ANPIYY63 pKa = 10.37HH64 pKa = 6.69AVSNFPYY71 pKa = 10.27YY72 pKa = 9.8PHH74 pKa = 6.53QRR76 pKa = 11.84LDD78 pKa = 2.94TSTFVPCTQVLFAVLAHH95 pKa = 6.78MDD97 pKa = 3.9NIMCTTYY104 pKa = 10.5SWTQSQPAWHH114 pKa = 6.94PCVSRR119 pKa = 11.84LYY121 pKa = 10.59FSTLVYY127 pKa = 9.96YY128 pKa = 10.22QILRR132 pKa = 11.84AMDD135 pKa = 4.09AARR138 pKa = 11.84IITVLQKK145 pKa = 10.76EE146 pKa = 4.26LLNFLEE152 pKa = 4.3HH153 pKa = 6.99TIPASALPIPGPLAPFLRR171 pKa = 11.84SPFLLFNWHH180 pKa = 6.01GWLLQCMSNHH190 pKa = 5.76TXWPHH195 pKa = 5.0HH196 pKa = 5.97QKK198 pKa = 9.99IKK200 pKa = 10.74YY201 pKa = 9.14NISSAFNNLLPNLVHH216 pKa = 6.95AVDD219 pKa = 3.7SAYY222 pKa = 11.3SFATDD227 pKa = 2.86ATYY230 pKa = 11.23VFRR233 pKa = 11.84SAVNDD238 pKa = 3.81LNVKK242 pKa = 9.39MEE244 pKa = 4.21ATDD247 pKa = 3.77PDD249 pKa = 3.69MPAAEE254 pKa = 4.07TRR256 pKa = 11.84HH257 pKa = 5.8WNSAMMDD264 pKa = 3.55PVYY267 pKa = 10.65RR268 pKa = 11.84NDD270 pKa = 3.03MYY272 pKa = 11.1IPTRR276 pKa = 11.84VAQNFAAFSAPLGLPLRR293 pKa = 11.84YY294 pKa = 9.81NYY296 pKa = 10.87GPDD299 pKa = 3.32TDD301 pKa = 4.0IGLSHH306 pKa = 7.55FLYY309 pKa = 10.87LSGGKK314 pKa = 9.0HH315 pKa = 7.0DD316 pKa = 4.29YY317 pKa = 11.07LNRR320 pKa = 11.84LAPVFARR327 pKa = 11.84YY328 pKa = 8.6SQYY331 pKa = 10.66FVNSKK336 pKa = 10.14SLQEE340 pKa = 4.04CSPVGSAPGQINGTLTDD357 pKa = 5.83AIPVPTISGTTVYY370 pKa = 10.22TPSTLAALVTPWSSTTIYY388 pKa = 9.47THH390 pKa = 6.58VNLSLPEE397 pKa = 4.07FFEE400 pKa = 4.38HH401 pKa = 6.5QAALTNVNVTEE412 pKa = 4.73HH413 pKa = 6.32GAAPTFGTTATTLNGSFWRR432 pKa = 11.84MKK434 pKa = 8.42PHH436 pKa = 5.8YY437 pKa = 9.82RR438 pKa = 11.84EE439 pKa = 4.18SPPINVTKK447 pKa = 10.8NLFNIISNEE456 pKa = 3.72MFAEE460 pKa = 4.44PTSS463 pKa = 3.57
Molecular weight: 51.33 kDa
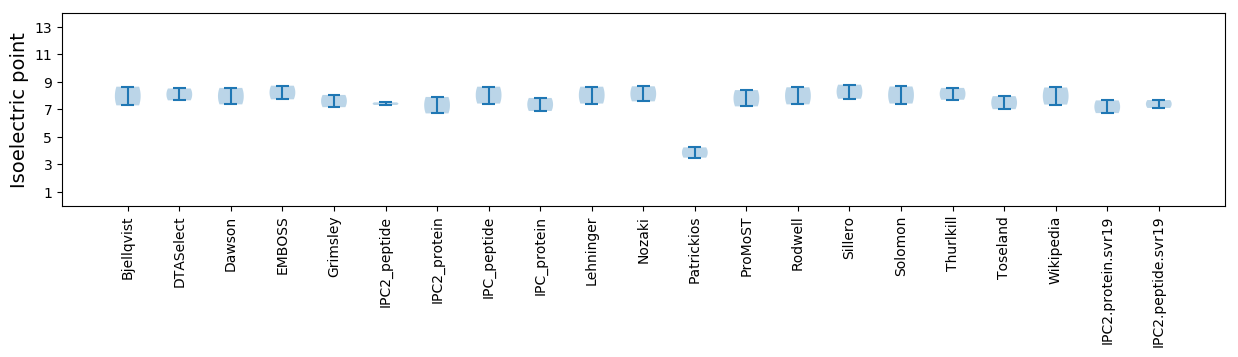
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0STM1|C0STM1_9VIRU Coat protein OS=Flammulina velutipes browning virus OX=568090 GN=CP1 PE=4 SV=1
MM1 pKa = 7.98SDD3 pKa = 3.18TLIDD7 pKa = 3.62SFSRR11 pKa = 11.84LTLSVKK17 pKa = 10.51NFVFLGFTEE26 pKa = 4.42TQNYY30 pKa = 6.17PQKK33 pKa = 10.49SDD35 pKa = 3.36SAILSHH41 pKa = 7.05RR42 pKa = 11.84KK43 pKa = 9.01VVLNAFEE50 pKa = 5.31KK51 pKa = 9.95YY52 pKa = 10.58LNPIEE57 pKa = 4.29YY58 pKa = 10.05NHH60 pKa = 6.39VANEE64 pKa = 4.12YY65 pKa = 10.18KK66 pKa = 10.51RR67 pKa = 11.84SEE69 pKa = 4.17TDD71 pKa = 3.2LDD73 pKa = 3.65STKK76 pKa = 11.05AAFFKK81 pKa = 11.1GDD83 pKa = 3.59IPDD86 pKa = 3.96HH87 pKa = 5.48EE88 pKa = 4.79VPRR91 pKa = 11.84DD92 pKa = 3.42EE93 pKa = 4.91HH94 pKa = 6.29YY95 pKa = 11.37NRR97 pKa = 11.84AFSVIVSKK105 pKa = 10.24FRR107 pKa = 11.84PPEE110 pKa = 4.18PIRR113 pKa = 11.84PVHH116 pKa = 5.64YY117 pKa = 10.57ADD119 pKa = 4.27LRR121 pKa = 11.84LYY123 pKa = 9.13PWPLKK128 pKa = 10.38PSAEE132 pKa = 4.1APFSNDD138 pKa = 2.51KK139 pKa = 11.13SLLALLALRR148 pKa = 11.84NRR150 pKa = 11.84QGFLPNAKK158 pKa = 9.95PNFHH162 pKa = 6.89NLFNWVFGFNRR173 pKa = 11.84QCVHH177 pKa = 7.27LIKK180 pKa = 10.45KK181 pKa = 10.36GKK183 pKa = 10.15DD184 pKa = 3.05NLGPNEE190 pKa = 4.09YY191 pKa = 9.46WPAHH195 pKa = 5.43GFLYY199 pKa = 9.93PINIHH204 pKa = 5.41TKK206 pKa = 8.42SAIIGIHH213 pKa = 6.96DD214 pKa = 4.12PNKK217 pKa = 8.91VRR219 pKa = 11.84TIFGVPKK226 pKa = 8.81LTVMVEE232 pKa = 4.99AMFFWPLFRR241 pKa = 11.84YY242 pKa = 9.79YY243 pKa = 10.41RR244 pKa = 11.84FEE246 pKa = 4.03QQSPLLWGYY255 pKa = 7.77EE256 pKa = 3.97TMLGGWYY263 pKa = 10.2KK264 pKa = 10.9LNHH267 pKa = 6.16EE268 pKa = 4.2LHH270 pKa = 6.88LNPFYY275 pKa = 10.62QGSILSLDD283 pKa = 3.0WSFFDD288 pKa = 4.21GRR290 pKa = 11.84ALFSVINDD298 pKa = 4.34LYY300 pKa = 10.78SDD302 pKa = 4.35KK303 pKa = 11.06GVKK306 pKa = 9.85SYY308 pKa = 11.62FEE310 pKa = 4.37FNNGYY315 pKa = 9.4IPTVDD320 pKa = 4.01YY321 pKa = 10.39PDD323 pKa = 4.81SSTHH327 pKa = 5.11PQKK330 pKa = 10.98LHH332 pKa = 6.56NLWDD336 pKa = 3.49WMLTALKK343 pKa = 9.73FAPCALADD351 pKa = 3.58GTIWQRR357 pKa = 11.84TVRR360 pKa = 11.84GIASGQFTTQFMDD373 pKa = 4.03SIYY376 pKa = 10.5NGLMILTILSRR387 pKa = 11.84MGFVIDD393 pKa = 3.33EE394 pKa = 4.23TLPIKK399 pKa = 10.76LLGDD403 pKa = 3.87DD404 pKa = 3.73SVTRR408 pKa = 11.84LAVSIPASMHH418 pKa = 5.53EE419 pKa = 4.1SFLIEE424 pKa = 4.04FQRR427 pKa = 11.84LADD430 pKa = 3.73YY431 pKa = 10.67YY432 pKa = 10.78FSHH435 pKa = 7.49TINVKK440 pKa = 9.87KK441 pKa = 10.7SKK443 pKa = 10.32ISNTPHH449 pKa = 5.76NVSVLSYY456 pKa = 10.93ANNNGLPVRR465 pKa = 11.84SRR467 pKa = 11.84TSLLCALLYY476 pKa = 9.77PKK478 pKa = 10.08SRR480 pKa = 11.84RR481 pKa = 11.84PTWEE485 pKa = 3.6HH486 pKa = 5.45LKK488 pKa = 10.75ARR490 pKa = 11.84AIGVYY495 pKa = 9.59YY496 pKa = 10.29ASCGIDD502 pKa = 2.83RR503 pKa = 11.84TVRR506 pKa = 11.84LICKK510 pKa = 10.15DD511 pKa = 2.74IFDD514 pKa = 4.53YY515 pKa = 11.25LDD517 pKa = 3.41SQGIQASSAGLQDD530 pKa = 5.16LFDD533 pKa = 5.61PNFKK537 pKa = 10.7SGTIPLDD544 pKa = 3.18VFPSVEE550 pKa = 3.99QVTTNLRR557 pKa = 11.84SFHH560 pKa = 6.83HH561 pKa = 7.06LDD563 pKa = 3.38NSDD566 pKa = 3.38KK567 pKa = 10.75EE568 pKa = 4.72RR569 pKa = 11.84YY570 pKa = 9.21FPTSHH575 pKa = 7.46FLDD578 pKa = 4.01TKK580 pKa = 11.01
MM1 pKa = 7.98SDD3 pKa = 3.18TLIDD7 pKa = 3.62SFSRR11 pKa = 11.84LTLSVKK17 pKa = 10.51NFVFLGFTEE26 pKa = 4.42TQNYY30 pKa = 6.17PQKK33 pKa = 10.49SDD35 pKa = 3.36SAILSHH41 pKa = 7.05RR42 pKa = 11.84KK43 pKa = 9.01VVLNAFEE50 pKa = 5.31KK51 pKa = 9.95YY52 pKa = 10.58LNPIEE57 pKa = 4.29YY58 pKa = 10.05NHH60 pKa = 6.39VANEE64 pKa = 4.12YY65 pKa = 10.18KK66 pKa = 10.51RR67 pKa = 11.84SEE69 pKa = 4.17TDD71 pKa = 3.2LDD73 pKa = 3.65STKK76 pKa = 11.05AAFFKK81 pKa = 11.1GDD83 pKa = 3.59IPDD86 pKa = 3.96HH87 pKa = 5.48EE88 pKa = 4.79VPRR91 pKa = 11.84DD92 pKa = 3.42EE93 pKa = 4.91HH94 pKa = 6.29YY95 pKa = 11.37NRR97 pKa = 11.84AFSVIVSKK105 pKa = 10.24FRR107 pKa = 11.84PPEE110 pKa = 4.18PIRR113 pKa = 11.84PVHH116 pKa = 5.64YY117 pKa = 10.57ADD119 pKa = 4.27LRR121 pKa = 11.84LYY123 pKa = 9.13PWPLKK128 pKa = 10.38PSAEE132 pKa = 4.1APFSNDD138 pKa = 2.51KK139 pKa = 11.13SLLALLALRR148 pKa = 11.84NRR150 pKa = 11.84QGFLPNAKK158 pKa = 9.95PNFHH162 pKa = 6.89NLFNWVFGFNRR173 pKa = 11.84QCVHH177 pKa = 7.27LIKK180 pKa = 10.45KK181 pKa = 10.36GKK183 pKa = 10.15DD184 pKa = 3.05NLGPNEE190 pKa = 4.09YY191 pKa = 9.46WPAHH195 pKa = 5.43GFLYY199 pKa = 9.93PINIHH204 pKa = 5.41TKK206 pKa = 8.42SAIIGIHH213 pKa = 6.96DD214 pKa = 4.12PNKK217 pKa = 8.91VRR219 pKa = 11.84TIFGVPKK226 pKa = 8.81LTVMVEE232 pKa = 4.99AMFFWPLFRR241 pKa = 11.84YY242 pKa = 9.79YY243 pKa = 10.41RR244 pKa = 11.84FEE246 pKa = 4.03QQSPLLWGYY255 pKa = 7.77EE256 pKa = 3.97TMLGGWYY263 pKa = 10.2KK264 pKa = 10.9LNHH267 pKa = 6.16EE268 pKa = 4.2LHH270 pKa = 6.88LNPFYY275 pKa = 10.62QGSILSLDD283 pKa = 3.0WSFFDD288 pKa = 4.21GRR290 pKa = 11.84ALFSVINDD298 pKa = 4.34LYY300 pKa = 10.78SDD302 pKa = 4.35KK303 pKa = 11.06GVKK306 pKa = 9.85SYY308 pKa = 11.62FEE310 pKa = 4.37FNNGYY315 pKa = 9.4IPTVDD320 pKa = 4.01YY321 pKa = 10.39PDD323 pKa = 4.81SSTHH327 pKa = 5.11PQKK330 pKa = 10.98LHH332 pKa = 6.56NLWDD336 pKa = 3.49WMLTALKK343 pKa = 9.73FAPCALADD351 pKa = 3.58GTIWQRR357 pKa = 11.84TVRR360 pKa = 11.84GIASGQFTTQFMDD373 pKa = 4.03SIYY376 pKa = 10.5NGLMILTILSRR387 pKa = 11.84MGFVIDD393 pKa = 3.33EE394 pKa = 4.23TLPIKK399 pKa = 10.76LLGDD403 pKa = 3.87DD404 pKa = 3.73SVTRR408 pKa = 11.84LAVSIPASMHH418 pKa = 5.53EE419 pKa = 4.1SFLIEE424 pKa = 4.04FQRR427 pKa = 11.84LADD430 pKa = 3.73YY431 pKa = 10.67YY432 pKa = 10.78FSHH435 pKa = 7.49TINVKK440 pKa = 9.87KK441 pKa = 10.7SKK443 pKa = 10.32ISNTPHH449 pKa = 5.76NVSVLSYY456 pKa = 10.93ANNNGLPVRR465 pKa = 11.84SRR467 pKa = 11.84TSLLCALLYY476 pKa = 9.77PKK478 pKa = 10.08SRR480 pKa = 11.84RR481 pKa = 11.84PTWEE485 pKa = 3.6HH486 pKa = 5.45LKK488 pKa = 10.75ARR490 pKa = 11.84AIGVYY495 pKa = 9.59YY496 pKa = 10.29ASCGIDD502 pKa = 2.83RR503 pKa = 11.84TVRR506 pKa = 11.84LICKK510 pKa = 10.15DD511 pKa = 2.74IFDD514 pKa = 4.53YY515 pKa = 11.25LDD517 pKa = 3.41SQGIQASSAGLQDD530 pKa = 5.16LFDD533 pKa = 5.61PNFKK537 pKa = 10.7SGTIPLDD544 pKa = 3.18VFPSVEE550 pKa = 3.99QVTTNLRR557 pKa = 11.84SFHH560 pKa = 6.83HH561 pKa = 7.06LDD563 pKa = 3.38NSDD566 pKa = 3.38KK567 pKa = 10.75EE568 pKa = 4.72RR569 pKa = 11.84YY570 pKa = 9.21FPTSHH575 pKa = 7.46FLDD578 pKa = 4.01TKK580 pKa = 11.01
Molecular weight: 66.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1043 |
463 |
580 |
521.5 |
59.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.999 ± 1.388 | 0.959 ± 0.081 |
5.081 ± 0.8 | 3.164 ± 0.383 |
6.136 ± 0.783 | 4.219 ± 0.511 |
3.643 ± 0.019 | 4.89 ± 0.671 |
4.219 ± 1.09 | 9.779 ± 0.764 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.013 ± 0.388 | 5.944 ± 0.069 |
7.862 ± 1.389 | 2.972 ± 0.179 |
4.506 ± 0.414 | 8.629 ± 0.296 |
6.711 ± 1.292 | 5.753 ± 0.342 |
1.822 ± 0.063 | 4.602 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
