
Barley yellow dwarf virus (isolate MAV-PS1) (BYDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus; Barley yellow dwarf virus MAV
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
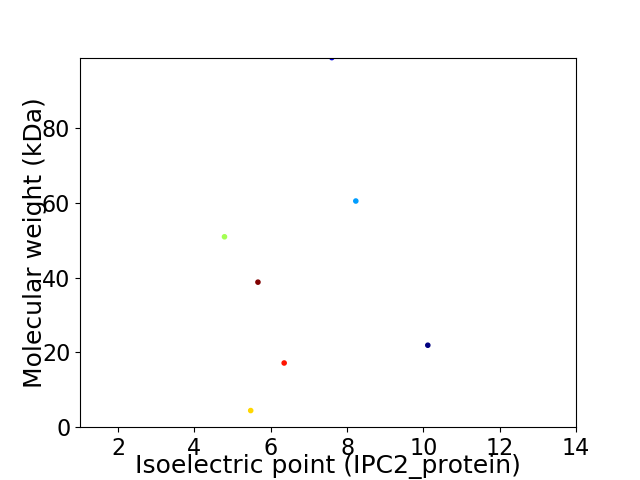
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65873|Q65873_BYDV1 ORF6 OS=Barley yellow dwarf virus (isolate MAV-PS1) OX=31723 PE=4 SV=1
VV1 pKa = 7.52DD2 pKa = 3.85SSTPEE7 pKa = 4.32PSPQPQPEE15 pKa = 4.54PKK17 pKa = 9.45PDD19 pKa = 3.89PQPTPEE25 pKa = 4.01PRR27 pKa = 11.84QKK29 pKa = 10.82RR30 pKa = 11.84FFEE33 pKa = 4.55YY34 pKa = 10.2VGTPYY39 pKa = 10.92VVIQTRR45 pKa = 11.84EE46 pKa = 3.99SSDD49 pKa = 3.82SIAVKK54 pKa = 10.7AMNDD58 pKa = 3.06QSFQYY63 pKa = 10.28IEE65 pKa = 4.51NEE67 pKa = 3.83TSEE70 pKa = 4.0QRR72 pKa = 11.84TVKK75 pKa = 10.49AWWNSNNSVQAQAAFIFPIPAGEE98 pKa = 3.91YY99 pKa = 9.93SVNISCEE106 pKa = 3.87GLQSVDD112 pKa = 3.65HH113 pKa = 6.79IGGNRR118 pKa = 11.84DD119 pKa = 3.66GYY121 pKa = 10.15WIGLIAYY128 pKa = 7.77QSQSGDD134 pKa = 3.12YY135 pKa = 8.92WGVGNYY141 pKa = 9.0VGCDD145 pKa = 2.86ITNLLGTNTWRR156 pKa = 11.84PGHH159 pKa = 6.74EE160 pKa = 3.96DD161 pKa = 4.64LEE163 pKa = 4.69LNSCKK168 pKa = 9.77FTDD171 pKa = 4.14GQIVEE176 pKa = 4.29RR177 pKa = 11.84DD178 pKa = 3.46AVISFHH184 pKa = 5.98VKK186 pKa = 10.21ARR188 pKa = 11.84GADD191 pKa = 3.29PKK193 pKa = 10.51FYY195 pKa = 11.18LMAPKK200 pKa = 9.75TMKK203 pKa = 10.13ADD205 pKa = 3.08KK206 pKa = 9.79YY207 pKa = 10.38NYY209 pKa = 7.68VVSYY213 pKa = 10.89GGYY216 pKa = 6.85TDD218 pKa = 3.8KK219 pKa = 11.13RR220 pKa = 11.84MEE222 pKa = 4.54FGTISVTVDD231 pKa = 3.07EE232 pKa = 5.17SDD234 pKa = 3.85VEE236 pKa = 4.04AEE238 pKa = 3.98RR239 pKa = 11.84YY240 pKa = 9.18SRR242 pKa = 11.84HH243 pKa = 5.04TSTVRR248 pKa = 11.84RR249 pKa = 11.84TEE251 pKa = 3.7NRR253 pKa = 11.84DD254 pKa = 3.64YY255 pKa = 11.54GWMNVLPPYY264 pKa = 10.8NPDD267 pKa = 3.07QVPEE271 pKa = 4.14QEE273 pKa = 4.76DD274 pKa = 3.79EE275 pKa = 4.27QPVVDD280 pKa = 4.82KK281 pKa = 11.25EE282 pKa = 3.98MDD284 pKa = 3.54AGSPIDD290 pKa = 3.9TASLTSDD297 pKa = 3.68TEE299 pKa = 4.03AEE301 pKa = 4.06KK302 pKa = 11.13AFDD305 pKa = 3.97LKK307 pKa = 11.2EE308 pKa = 4.16EE309 pKa = 4.11EE310 pKa = 4.09LTRR313 pKa = 11.84AILEE317 pKa = 4.26YY318 pKa = 10.43EE319 pKa = 4.18AATVSIPDD327 pKa = 3.99AAPDD331 pKa = 3.52ILPSKK336 pKa = 10.88SEE338 pKa = 4.0MSSKK342 pKa = 10.68PIDD345 pKa = 3.54RR346 pKa = 11.84DD347 pKa = 3.49GRR349 pKa = 11.84SLPKK353 pKa = 10.44SQTKK357 pKa = 9.47EE358 pKa = 3.7VLGTYY363 pKa = 9.39QGQNITSDD371 pKa = 3.62DD372 pKa = 4.18VPPVIAEE379 pKa = 4.08KK380 pKa = 10.58LRR382 pKa = 11.84EE383 pKa = 4.08VNRR386 pKa = 11.84APSTLLYY393 pKa = 10.5DD394 pKa = 4.33RR395 pKa = 11.84QPKK398 pKa = 8.56QPKK401 pKa = 9.36NPLTRR406 pKa = 11.84FVLSNKK412 pKa = 9.13TSTASPGSQSSTAGMTRR429 pKa = 11.84EE430 pKa = 3.89QASEE434 pKa = 3.75YY435 pKa = 9.62TRR437 pKa = 11.84IRR439 pKa = 11.84KK440 pKa = 9.16SLGLTAAKK448 pKa = 9.89QYY450 pKa = 10.54KK451 pKa = 10.32ASLDD455 pKa = 3.68DD456 pKa = 3.54TT457 pKa = 4.51
VV1 pKa = 7.52DD2 pKa = 3.85SSTPEE7 pKa = 4.32PSPQPQPEE15 pKa = 4.54PKK17 pKa = 9.45PDD19 pKa = 3.89PQPTPEE25 pKa = 4.01PRR27 pKa = 11.84QKK29 pKa = 10.82RR30 pKa = 11.84FFEE33 pKa = 4.55YY34 pKa = 10.2VGTPYY39 pKa = 10.92VVIQTRR45 pKa = 11.84EE46 pKa = 3.99SSDD49 pKa = 3.82SIAVKK54 pKa = 10.7AMNDD58 pKa = 3.06QSFQYY63 pKa = 10.28IEE65 pKa = 4.51NEE67 pKa = 3.83TSEE70 pKa = 4.0QRR72 pKa = 11.84TVKK75 pKa = 10.49AWWNSNNSVQAQAAFIFPIPAGEE98 pKa = 3.91YY99 pKa = 9.93SVNISCEE106 pKa = 3.87GLQSVDD112 pKa = 3.65HH113 pKa = 6.79IGGNRR118 pKa = 11.84DD119 pKa = 3.66GYY121 pKa = 10.15WIGLIAYY128 pKa = 7.77QSQSGDD134 pKa = 3.12YY135 pKa = 8.92WGVGNYY141 pKa = 9.0VGCDD145 pKa = 2.86ITNLLGTNTWRR156 pKa = 11.84PGHH159 pKa = 6.74EE160 pKa = 3.96DD161 pKa = 4.64LEE163 pKa = 4.69LNSCKK168 pKa = 9.77FTDD171 pKa = 4.14GQIVEE176 pKa = 4.29RR177 pKa = 11.84DD178 pKa = 3.46AVISFHH184 pKa = 5.98VKK186 pKa = 10.21ARR188 pKa = 11.84GADD191 pKa = 3.29PKK193 pKa = 10.51FYY195 pKa = 11.18LMAPKK200 pKa = 9.75TMKK203 pKa = 10.13ADD205 pKa = 3.08KK206 pKa = 9.79YY207 pKa = 10.38NYY209 pKa = 7.68VVSYY213 pKa = 10.89GGYY216 pKa = 6.85TDD218 pKa = 3.8KK219 pKa = 11.13RR220 pKa = 11.84MEE222 pKa = 4.54FGTISVTVDD231 pKa = 3.07EE232 pKa = 5.17SDD234 pKa = 3.85VEE236 pKa = 4.04AEE238 pKa = 3.98RR239 pKa = 11.84YY240 pKa = 9.18SRR242 pKa = 11.84HH243 pKa = 5.04TSTVRR248 pKa = 11.84RR249 pKa = 11.84TEE251 pKa = 3.7NRR253 pKa = 11.84DD254 pKa = 3.64YY255 pKa = 11.54GWMNVLPPYY264 pKa = 10.8NPDD267 pKa = 3.07QVPEE271 pKa = 4.14QEE273 pKa = 4.76DD274 pKa = 3.79EE275 pKa = 4.27QPVVDD280 pKa = 4.82KK281 pKa = 11.25EE282 pKa = 3.98MDD284 pKa = 3.54AGSPIDD290 pKa = 3.9TASLTSDD297 pKa = 3.68TEE299 pKa = 4.03AEE301 pKa = 4.06KK302 pKa = 11.13AFDD305 pKa = 3.97LKK307 pKa = 11.2EE308 pKa = 4.16EE309 pKa = 4.11EE310 pKa = 4.09LTRR313 pKa = 11.84AILEE317 pKa = 4.26YY318 pKa = 10.43EE319 pKa = 4.18AATVSIPDD327 pKa = 3.99AAPDD331 pKa = 3.52ILPSKK336 pKa = 10.88SEE338 pKa = 4.0MSSKK342 pKa = 10.68PIDD345 pKa = 3.54RR346 pKa = 11.84DD347 pKa = 3.49GRR349 pKa = 11.84SLPKK353 pKa = 10.44SQTKK357 pKa = 9.47EE358 pKa = 3.7VLGTYY363 pKa = 9.39QGQNITSDD371 pKa = 3.62DD372 pKa = 4.18VPPVIAEE379 pKa = 4.08KK380 pKa = 10.58LRR382 pKa = 11.84EE383 pKa = 4.08VNRR386 pKa = 11.84APSTLLYY393 pKa = 10.5DD394 pKa = 4.33RR395 pKa = 11.84QPKK398 pKa = 8.56QPKK401 pKa = 9.36NPLTRR406 pKa = 11.84FVLSNKK412 pKa = 9.13TSTASPGSQSSTAGMTRR429 pKa = 11.84EE430 pKa = 3.89QASEE434 pKa = 3.75YY435 pKa = 9.62TRR437 pKa = 11.84IRR439 pKa = 11.84KK440 pKa = 9.16SLGLTAAKK448 pKa = 9.89QYY450 pKa = 10.54KK451 pKa = 10.32ASLDD455 pKa = 3.68DD456 pKa = 3.54TT457 pKa = 4.51
Molecular weight: 50.95 kDa
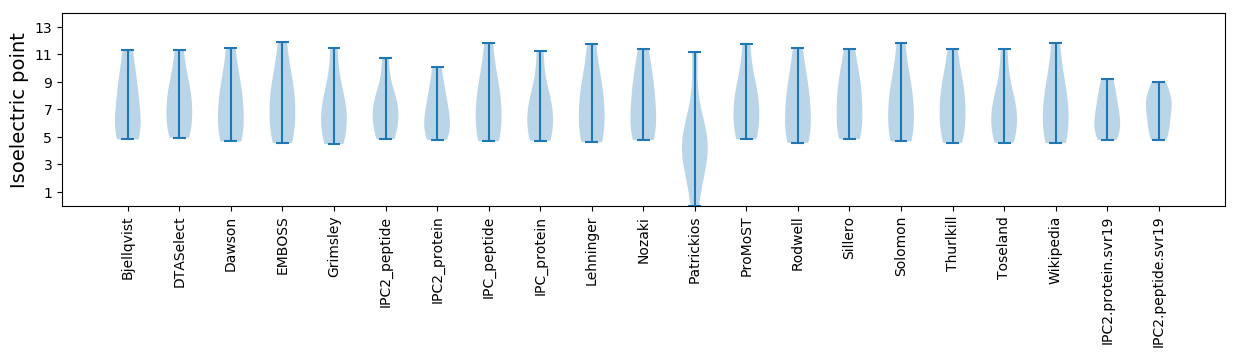
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65871|Q65871_BYDV1 RNA-directed RNA polymerase (Fragment) OS=Barley yellow dwarf virus (isolate MAV-PS1) OX=31723 PE=4 SV=1
MM1 pKa = 7.28NSVGRR6 pKa = 11.84RR7 pKa = 11.84NNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84NGPRR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84VSAVRR25 pKa = 11.84RR26 pKa = 11.84MVVVQPNRR34 pKa = 11.84AGPKK38 pKa = 8.16RR39 pKa = 11.84RR40 pKa = 11.84ARR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84TRR46 pKa = 11.84GGGANLISGPAGRR59 pKa = 11.84TEE61 pKa = 3.81VFVFSVNDD69 pKa = 3.76LKK71 pKa = 11.54ANSSGTIKK79 pKa = 10.68FGPDD83 pKa = 3.19LSQCPALSGGILKK96 pKa = 10.33SYY98 pKa = 10.55HH99 pKa = 6.6LYY101 pKa = 10.64KK102 pKa = 9.46ITNVKK107 pKa = 10.26VEE109 pKa = 4.92FKK111 pKa = 10.37SHH113 pKa = 6.16ASASTVGAMFIEE125 pKa = 5.12LDD127 pKa = 3.67TWCSQSTLGSYY138 pKa = 10.37INSFTISKK146 pKa = 10.2SATKK150 pKa = 10.09TFTAQQIDD158 pKa = 3.42GKK160 pKa = 7.87EE161 pKa = 4.04FRR163 pKa = 11.84EE164 pKa = 4.2STVNQFYY171 pKa = 10.29MLYY174 pKa = 9.29KK175 pKa = 10.81ANGSTSDD182 pKa = 3.18TAGQFIITIRR192 pKa = 11.84VANMTPKK199 pKa = 10.68
MM1 pKa = 7.28NSVGRR6 pKa = 11.84RR7 pKa = 11.84NNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84NGPRR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84VSAVRR25 pKa = 11.84RR26 pKa = 11.84MVVVQPNRR34 pKa = 11.84AGPKK38 pKa = 8.16RR39 pKa = 11.84RR40 pKa = 11.84ARR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84TRR46 pKa = 11.84GGGANLISGPAGRR59 pKa = 11.84TEE61 pKa = 3.81VFVFSVNDD69 pKa = 3.76LKK71 pKa = 11.54ANSSGTIKK79 pKa = 10.68FGPDD83 pKa = 3.19LSQCPALSGGILKK96 pKa = 10.33SYY98 pKa = 10.55HH99 pKa = 6.6LYY101 pKa = 10.64KK102 pKa = 9.46ITNVKK107 pKa = 10.26VEE109 pKa = 4.92FKK111 pKa = 10.37SHH113 pKa = 6.16ASASTVGAMFIEE125 pKa = 5.12LDD127 pKa = 3.67TWCSQSTLGSYY138 pKa = 10.37INSFTISKK146 pKa = 10.2SATKK150 pKa = 10.09TFTAQQIDD158 pKa = 3.42GKK160 pKa = 7.87EE161 pKa = 4.04FRR163 pKa = 11.84EE164 pKa = 4.2STVNQFYY171 pKa = 10.29MLYY174 pKa = 9.29KK175 pKa = 10.81ANGSTSDD182 pKa = 3.18TAGQFIITIRR192 pKa = 11.84VANMTPKK199 pKa = 10.68
Molecular weight: 21.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2591 |
43 |
867 |
370.1 |
41.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.294 ± 0.458 | 1.621 ± 0.223 |
5.326 ± 0.541 | 7.102 ± 0.881 |
4.091 ± 0.396 | 5.712 ± 0.434 |
2.084 ± 0.354 | 5.403 ± 0.457 |
7.294 ± 0.816 | 7.487 ± 0.565 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.663 ± 0.279 | 4.207 ± 0.363 |
4.323 ± 0.832 | 4.245 ± 0.77 |
5.866 ± 0.585 | 7.951 ± 0.605 |
5.712 ± 0.609 | 6.677 ± 0.323 |
1.003 ± 0.124 | 3.937 ± 0.406 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
