
Human papillomavirus 24
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 1
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
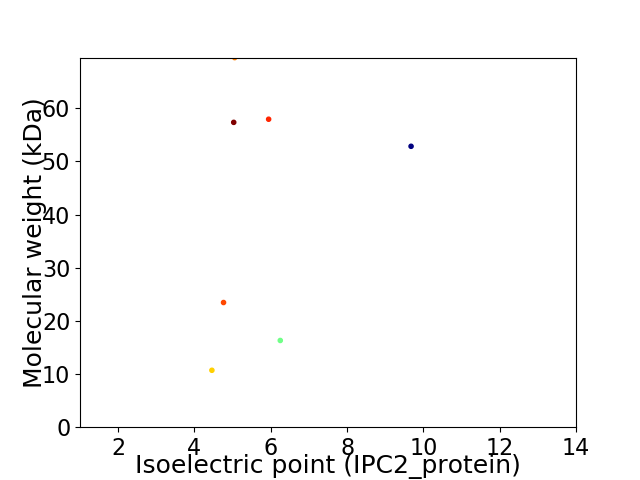
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
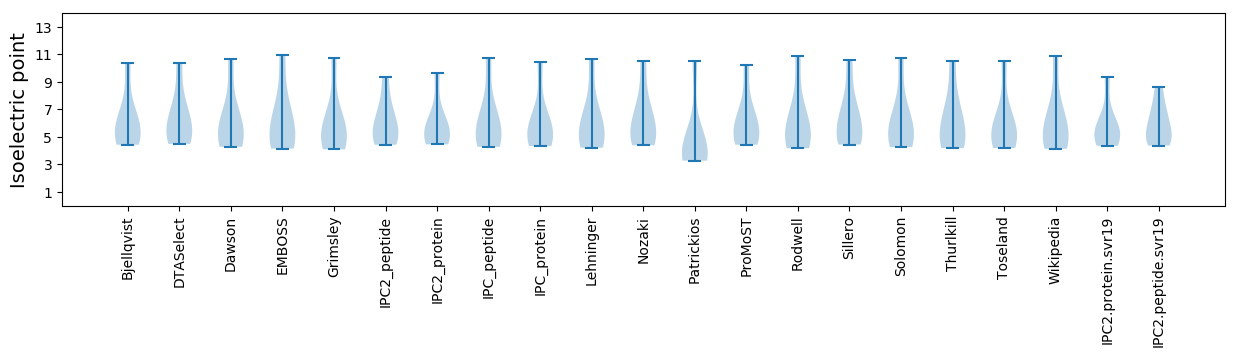
* You can choose from 21 different methods for calculating isoelectric point
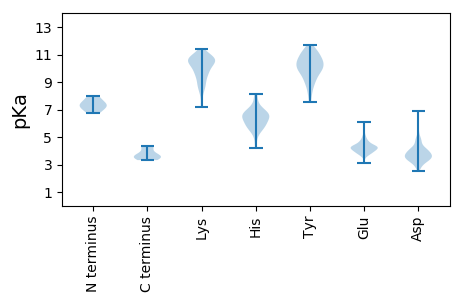
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P50790|VL1_HPV24 Major capsid protein L1 OS=Human papillomavirus 24 OX=37956 GN=L1 PE=3 SV=1
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.06IVLEE14 pKa = 4.09LTEE17 pKa = 4.4PQTVDD22 pKa = 2.9LHH24 pKa = 8.11CEE26 pKa = 4.01EE27 pKa = 5.16EE28 pKa = 4.64LPEE31 pKa = 4.02QDD33 pKa = 3.61TEE35 pKa = 4.39VEE37 pKa = 4.2PEE39 pKa = 3.43RR40 pKa = 11.84RR41 pKa = 11.84AYY43 pKa = 9.56KK44 pKa = 10.26IILCCGGGCGTRR56 pKa = 11.84LRR58 pKa = 11.84LFVAATQFGIRR69 pKa = 11.84GLQDD73 pKa = 3.58LLLEE77 pKa = 4.39EE78 pKa = 4.93VVILCPDD85 pKa = 3.54CRR87 pKa = 11.84NSDD90 pKa = 3.89LQHH93 pKa = 6.61GGQQ96 pKa = 3.6
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.06IVLEE14 pKa = 4.09LTEE17 pKa = 4.4PQTVDD22 pKa = 2.9LHH24 pKa = 8.11CEE26 pKa = 4.01EE27 pKa = 5.16EE28 pKa = 4.64LPEE31 pKa = 4.02QDD33 pKa = 3.61TEE35 pKa = 4.39VEE37 pKa = 4.2PEE39 pKa = 3.43RR40 pKa = 11.84RR41 pKa = 11.84AYY43 pKa = 9.56KK44 pKa = 10.26IILCCGGGCGTRR56 pKa = 11.84LRR58 pKa = 11.84LFVAATQFGIRR69 pKa = 11.84GLQDD73 pKa = 3.58LLLEE77 pKa = 4.39EE78 pKa = 4.93VVILCPDD85 pKa = 3.54CRR87 pKa = 11.84NSDD90 pKa = 3.89LQHH93 pKa = 6.61GGQQ96 pKa = 3.6
Molecular weight: 10.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P50777|VE6_HPV24 Protein E6 OS=Human papillomavirus 24 OX=37956 GN=E6 PE=3 SV=1
MM1 pKa = 7.68EE2 pKa = 4.75NLKK5 pKa = 10.64KK6 pKa = 10.63RR7 pKa = 11.84FDD9 pKa = 3.8VLQDD13 pKa = 3.53LLMNIYY19 pKa = 9.79EE20 pKa = 4.28QGSDD24 pKa = 3.13TLEE27 pKa = 3.98SQIEE31 pKa = 3.96HH32 pKa = 5.42WQALRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.31AVLLYY44 pKa = 9.22YY45 pKa = 10.62ARR47 pKa = 11.84QNGVLRR53 pKa = 11.84LGYY56 pKa = 10.65LPVPPLATSEE66 pKa = 4.54AKK68 pKa = 10.21AKK70 pKa = 10.05QAISMVLQLQSLQQSPYY87 pKa = 9.25GTEE90 pKa = 3.14KK91 pKa = 8.53WTLVDD96 pKa = 3.44TSIEE100 pKa = 4.32TFKK103 pKa = 10.35NTPEE107 pKa = 3.73NHH109 pKa = 6.18FKK111 pKa = 10.72KK112 pKa = 10.82GPINVEE118 pKa = 3.94VIYY121 pKa = 11.13DD122 pKa = 4.16GDD124 pKa = 4.07PDD126 pKa = 3.81NANLYY131 pKa = 8.07TMWKK135 pKa = 7.96YY136 pKa = 11.05VYY138 pKa = 11.28YY139 pKa = 10.15MDD141 pKa = 6.88DD142 pKa = 3.2NDD144 pKa = 3.31QWQKK148 pKa = 9.6TEE150 pKa = 3.72SGANHH155 pKa = 6.05TGIYY159 pKa = 9.67YY160 pKa = 10.58LIGEE164 pKa = 4.75FKK166 pKa = 10.36HH167 pKa = 6.48YY168 pKa = 10.62YY169 pKa = 10.29VLFADD174 pKa = 4.16DD175 pKa = 3.92ANRR178 pKa = 11.84YY179 pKa = 8.46SKK181 pKa = 10.61SGQWEE186 pKa = 3.78VRR188 pKa = 11.84INKK191 pKa = 7.16EE192 pKa = 4.11TVFAPVTSSTPPDD205 pKa = 3.48SPGGSRR211 pKa = 11.84EE212 pKa = 4.24LPGSTANSKK221 pKa = 10.66ASSPTQQPQQACSDD235 pKa = 3.65EE236 pKa = 4.2TTKK239 pKa = 10.62RR240 pKa = 11.84KK241 pKa = 9.97RR242 pKa = 11.84YY243 pKa = 8.9GRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 3.88SSPTDD252 pKa = 3.09SRR254 pKa = 11.84CRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84SSSRR262 pKa = 11.84QKK264 pKa = 10.04KK265 pKa = 7.31QGRR268 pKa = 11.84RR269 pKa = 11.84ARR271 pKa = 11.84SRR273 pKa = 11.84TRR275 pKa = 11.84SRR277 pKa = 11.84CSSTQTRR284 pKa = 11.84SRR286 pKa = 11.84STSRR290 pKa = 11.84RR291 pKa = 11.84SRR293 pKa = 11.84STSRR297 pKa = 11.84GNRR300 pKa = 11.84RR301 pKa = 11.84CRR303 pKa = 11.84GDD305 pKa = 3.24TPRR308 pKa = 11.84GQRR311 pKa = 11.84GVSTSSRR318 pKa = 11.84GRR320 pKa = 11.84GRR322 pKa = 11.84GSRR325 pKa = 11.84RR326 pKa = 11.84SSSSSSPTPRR336 pKa = 11.84TKK338 pKa = 10.67ASQRR342 pKa = 11.84GCDD345 pKa = 3.33TRR347 pKa = 11.84SVRR350 pKa = 11.84DD351 pKa = 3.59SGISPGDD358 pKa = 3.42VGRR361 pKa = 11.84KK362 pKa = 8.3LQTVSGRR369 pKa = 11.84NSGRR373 pKa = 11.84LGRR376 pKa = 11.84LLEE379 pKa = 4.34EE380 pKa = 4.6ALDD383 pKa = 3.9PPVILLRR390 pKa = 11.84GGANTLKK397 pKa = 10.56CFRR400 pKa = 11.84NRR402 pKa = 11.84AKK404 pKa = 10.47LRR406 pKa = 11.84YY407 pKa = 8.28RR408 pKa = 11.84GHH410 pKa = 6.07YY411 pKa = 9.54KK412 pKa = 10.53AFSTSWSWVAADD424 pKa = 3.35GTEE427 pKa = 3.74RR428 pKa = 11.84LGRR431 pKa = 11.84SRR433 pKa = 11.84LLVSFTSFKK442 pKa = 10.4QRR444 pKa = 11.84SGFLDD449 pKa = 3.59LVRR452 pKa = 11.84FPKK455 pKa = 10.67GVDD458 pKa = 2.85WSLGSFDD465 pKa = 4.85KK466 pKa = 11.38LL467 pKa = 3.56
MM1 pKa = 7.68EE2 pKa = 4.75NLKK5 pKa = 10.64KK6 pKa = 10.63RR7 pKa = 11.84FDD9 pKa = 3.8VLQDD13 pKa = 3.53LLMNIYY19 pKa = 9.79EE20 pKa = 4.28QGSDD24 pKa = 3.13TLEE27 pKa = 3.98SQIEE31 pKa = 3.96HH32 pKa = 5.42WQALRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.31AVLLYY44 pKa = 9.22YY45 pKa = 10.62ARR47 pKa = 11.84QNGVLRR53 pKa = 11.84LGYY56 pKa = 10.65LPVPPLATSEE66 pKa = 4.54AKK68 pKa = 10.21AKK70 pKa = 10.05QAISMVLQLQSLQQSPYY87 pKa = 9.25GTEE90 pKa = 3.14KK91 pKa = 8.53WTLVDD96 pKa = 3.44TSIEE100 pKa = 4.32TFKK103 pKa = 10.35NTPEE107 pKa = 3.73NHH109 pKa = 6.18FKK111 pKa = 10.72KK112 pKa = 10.82GPINVEE118 pKa = 3.94VIYY121 pKa = 11.13DD122 pKa = 4.16GDD124 pKa = 4.07PDD126 pKa = 3.81NANLYY131 pKa = 8.07TMWKK135 pKa = 7.96YY136 pKa = 11.05VYY138 pKa = 11.28YY139 pKa = 10.15MDD141 pKa = 6.88DD142 pKa = 3.2NDD144 pKa = 3.31QWQKK148 pKa = 9.6TEE150 pKa = 3.72SGANHH155 pKa = 6.05TGIYY159 pKa = 9.67YY160 pKa = 10.58LIGEE164 pKa = 4.75FKK166 pKa = 10.36HH167 pKa = 6.48YY168 pKa = 10.62YY169 pKa = 10.29VLFADD174 pKa = 4.16DD175 pKa = 3.92ANRR178 pKa = 11.84YY179 pKa = 8.46SKK181 pKa = 10.61SGQWEE186 pKa = 3.78VRR188 pKa = 11.84INKK191 pKa = 7.16EE192 pKa = 4.11TVFAPVTSSTPPDD205 pKa = 3.48SPGGSRR211 pKa = 11.84EE212 pKa = 4.24LPGSTANSKK221 pKa = 10.66ASSPTQQPQQACSDD235 pKa = 3.65EE236 pKa = 4.2TTKK239 pKa = 10.62RR240 pKa = 11.84KK241 pKa = 9.97RR242 pKa = 11.84YY243 pKa = 8.9GRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 3.88SSPTDD252 pKa = 3.09SRR254 pKa = 11.84CRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84SSSRR262 pKa = 11.84QKK264 pKa = 10.04KK265 pKa = 7.31QGRR268 pKa = 11.84RR269 pKa = 11.84ARR271 pKa = 11.84SRR273 pKa = 11.84TRR275 pKa = 11.84SRR277 pKa = 11.84CSSTQTRR284 pKa = 11.84SRR286 pKa = 11.84STSRR290 pKa = 11.84RR291 pKa = 11.84SRR293 pKa = 11.84STSRR297 pKa = 11.84GNRR300 pKa = 11.84RR301 pKa = 11.84CRR303 pKa = 11.84GDD305 pKa = 3.24TPRR308 pKa = 11.84GQRR311 pKa = 11.84GVSTSSRR318 pKa = 11.84GRR320 pKa = 11.84GRR322 pKa = 11.84GSRR325 pKa = 11.84RR326 pKa = 11.84SSSSSSPTPRR336 pKa = 11.84TKK338 pKa = 10.67ASQRR342 pKa = 11.84GCDD345 pKa = 3.33TRR347 pKa = 11.84SVRR350 pKa = 11.84DD351 pKa = 3.59SGISPGDD358 pKa = 3.42VGRR361 pKa = 11.84KK362 pKa = 8.3LQTVSGRR369 pKa = 11.84NSGRR373 pKa = 11.84LGRR376 pKa = 11.84LLEE379 pKa = 4.34EE380 pKa = 4.6ALDD383 pKa = 3.9PPVILLRR390 pKa = 11.84GGANTLKK397 pKa = 10.56CFRR400 pKa = 11.84NRR402 pKa = 11.84AKK404 pKa = 10.47LRR406 pKa = 11.84YY407 pKa = 8.28RR408 pKa = 11.84GHH410 pKa = 6.07YY411 pKa = 9.54KK412 pKa = 10.53AFSTSWSWVAADD424 pKa = 3.35GTEE427 pKa = 3.74RR428 pKa = 11.84LGRR431 pKa = 11.84SRR433 pKa = 11.84LLVSFTSFKK442 pKa = 10.4QRR444 pKa = 11.84SGFLDD449 pKa = 3.59LVRR452 pKa = 11.84FPKK455 pKa = 10.67GVDD458 pKa = 2.85WSLGSFDD465 pKa = 4.85KK466 pKa = 11.38LL467 pKa = 3.56
Molecular weight: 52.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2568 |
96 |
607 |
366.9 |
41.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.607 ± 0.384 | 2.064 ± 0.659 |
5.919 ± 0.237 | 6.659 ± 0.723 |
3.933 ± 0.628 | 6.737 ± 1.044 |
1.908 ± 0.272 | 4.4 ± 0.566 |
4.673 ± 0.779 | 9.034 ± 0.777 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.363 ± 0.383 | 4.361 ± 0.646 |
6.62 ± 1.79 | 5.023 ± 0.191 |
6.737 ± 1.35 | 8.255 ± 1.031 |
6.347 ± 0.642 | 5.841 ± 0.673 |
1.285 ± 0.378 | 3.232 ± 0.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
