
Streptomyces sp. MP131-18
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
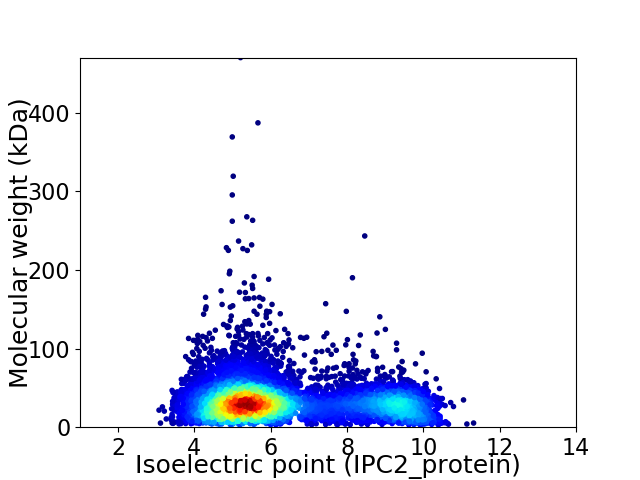
Virtual 2D-PAGE plot for 7037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V2RL99|A0A1V2RL99_9ACTN Uncharacterized protein OS=Streptomyces sp. MP131-18 OX=1857892 GN=STBA_39260 PE=4 SV=1
MM1 pKa = 7.45RR2 pKa = 11.84TTGRR6 pKa = 11.84IALAVLTATTLLLTACSSNDD26 pKa = 3.44RR27 pKa = 11.84PDD29 pKa = 3.35EE30 pKa = 4.33RR31 pKa = 11.84SPSDD35 pKa = 3.72PEE37 pKa = 4.26SDD39 pKa = 3.51ASSPAPDD46 pKa = 3.75APALEE51 pKa = 4.61PLPEE55 pKa = 4.71EE56 pKa = 4.72IPAEE60 pKa = 3.88LQPYY64 pKa = 8.77YY65 pKa = 10.16EE66 pKa = 5.44QEE68 pKa = 4.4LTWSSCGAVGFQCATLAVPLDD89 pKa = 4.0YY90 pKa = 11.51EE91 pKa = 4.47NVDD94 pKa = 3.6PAQDD98 pKa = 3.18IQLTVTRR105 pKa = 11.84ARR107 pKa = 11.84ATEE110 pKa = 3.78SGEE113 pKa = 4.17RR114 pKa = 11.84IGALLMNPGGPGASAVDD131 pKa = 4.38FAQDD135 pKa = 2.94AATYY139 pKa = 9.0LYY141 pKa = 9.62PPEE144 pKa = 3.75VRR146 pKa = 11.84ARR148 pKa = 11.84YY149 pKa = 10.6DD150 pKa = 3.33MVGLDD155 pKa = 4.25ARR157 pKa = 11.84GTGQSEE163 pKa = 4.08PVEE166 pKa = 4.59CLTGDD171 pKa = 3.91AMDD174 pKa = 6.05DD175 pKa = 3.56YY176 pKa = 10.72TLTDD180 pKa = 3.7RR181 pKa = 11.84TPDD184 pKa = 2.86SDD186 pKa = 4.85AEE188 pKa = 4.12VDD190 pKa = 3.94EE191 pKa = 5.47LLAAYY196 pKa = 10.3DD197 pKa = 5.37DD198 pKa = 4.41FAQGCQEE205 pKa = 3.71RR206 pKa = 11.84SGALLEE212 pKa = 4.95HH213 pKa = 7.06ISTIEE218 pKa = 3.9SARR221 pKa = 11.84DD222 pKa = 3.37MDD224 pKa = 4.29VLRR227 pKa = 11.84AALGDD232 pKa = 3.83EE233 pKa = 3.99KK234 pKa = 11.06LHH236 pKa = 5.82YY237 pKa = 10.88VGFSYY242 pKa = 8.92GTKK245 pKa = 10.42LGATYY250 pKa = 10.81AGLFPQRR257 pKa = 11.84TGRR260 pKa = 11.84LVLDD264 pKa = 3.95AAMDD268 pKa = 4.08PRR270 pKa = 11.84LSTLDD275 pKa = 3.35TDD277 pKa = 4.37RR278 pKa = 11.84EE279 pKa = 4.1QAGGFEE285 pKa = 4.07TAFRR289 pKa = 11.84SFAEE293 pKa = 4.38DD294 pKa = 3.26CTPRR298 pKa = 11.84DD299 pKa = 3.45SCALGNDD306 pKa = 4.17GADD309 pKa = 3.7GASQALLDD317 pKa = 4.49FFAEE321 pKa = 4.05VDD323 pKa = 3.92AEE325 pKa = 4.09PLPTGDD331 pKa = 3.71EE332 pKa = 4.09EE333 pKa = 5.36RR334 pKa = 11.84PLTEE338 pKa = 4.23SLASTGVSYY347 pKa = 11.45ALYY350 pKa = 10.8AEE352 pKa = 5.14DD353 pKa = 4.28LWPMLRR359 pKa = 11.84DD360 pKa = 3.64ALATAIEE367 pKa = 4.94DD368 pKa = 3.94GDD370 pKa = 4.13GSALLDD376 pKa = 4.1LADD379 pKa = 4.61AYY381 pKa = 11.05NEE383 pKa = 4.08RR384 pKa = 11.84EE385 pKa = 4.12ANGTYY390 pKa = 8.32GTTMFAFPAISCLDD404 pKa = 3.61SPAGNEE410 pKa = 3.87DD411 pKa = 3.31ADD413 pKa = 3.99AVRR416 pKa = 11.84SALAAYY422 pKa = 9.37EE423 pKa = 4.11EE424 pKa = 4.49ASPTFGRR431 pKa = 11.84DD432 pKa = 3.42FAWATLLCAAWPVDD446 pKa = 3.61PSGAPVSIPAADD458 pKa = 3.45ATDD461 pKa = 3.22ILVIGTLRR469 pKa = 11.84DD470 pKa = 3.29PATPYY475 pKa = 10.98AWAEE479 pKa = 3.9GLADD483 pKa = 3.68QLEE486 pKa = 4.49TGILLTYY493 pKa = 10.73DD494 pKa = 3.68GDD496 pKa = 3.74GHH498 pKa = 6.1GAYY501 pKa = 9.86GGASEE506 pKa = 5.15CVDD509 pKa = 3.56SAVNDD514 pKa = 3.69YY515 pKa = 11.17LLEE518 pKa = 4.39NVSPEE523 pKa = 4.96DD524 pKa = 3.5GTTCC528 pKa = 4.71
MM1 pKa = 7.45RR2 pKa = 11.84TTGRR6 pKa = 11.84IALAVLTATTLLLTACSSNDD26 pKa = 3.44RR27 pKa = 11.84PDD29 pKa = 3.35EE30 pKa = 4.33RR31 pKa = 11.84SPSDD35 pKa = 3.72PEE37 pKa = 4.26SDD39 pKa = 3.51ASSPAPDD46 pKa = 3.75APALEE51 pKa = 4.61PLPEE55 pKa = 4.71EE56 pKa = 4.72IPAEE60 pKa = 3.88LQPYY64 pKa = 8.77YY65 pKa = 10.16EE66 pKa = 5.44QEE68 pKa = 4.4LTWSSCGAVGFQCATLAVPLDD89 pKa = 4.0YY90 pKa = 11.51EE91 pKa = 4.47NVDD94 pKa = 3.6PAQDD98 pKa = 3.18IQLTVTRR105 pKa = 11.84ARR107 pKa = 11.84ATEE110 pKa = 3.78SGEE113 pKa = 4.17RR114 pKa = 11.84IGALLMNPGGPGASAVDD131 pKa = 4.38FAQDD135 pKa = 2.94AATYY139 pKa = 9.0LYY141 pKa = 9.62PPEE144 pKa = 3.75VRR146 pKa = 11.84ARR148 pKa = 11.84YY149 pKa = 10.6DD150 pKa = 3.33MVGLDD155 pKa = 4.25ARR157 pKa = 11.84GTGQSEE163 pKa = 4.08PVEE166 pKa = 4.59CLTGDD171 pKa = 3.91AMDD174 pKa = 6.05DD175 pKa = 3.56YY176 pKa = 10.72TLTDD180 pKa = 3.7RR181 pKa = 11.84TPDD184 pKa = 2.86SDD186 pKa = 4.85AEE188 pKa = 4.12VDD190 pKa = 3.94EE191 pKa = 5.47LLAAYY196 pKa = 10.3DD197 pKa = 5.37DD198 pKa = 4.41FAQGCQEE205 pKa = 3.71RR206 pKa = 11.84SGALLEE212 pKa = 4.95HH213 pKa = 7.06ISTIEE218 pKa = 3.9SARR221 pKa = 11.84DD222 pKa = 3.37MDD224 pKa = 4.29VLRR227 pKa = 11.84AALGDD232 pKa = 3.83EE233 pKa = 3.99KK234 pKa = 11.06LHH236 pKa = 5.82YY237 pKa = 10.88VGFSYY242 pKa = 8.92GTKK245 pKa = 10.42LGATYY250 pKa = 10.81AGLFPQRR257 pKa = 11.84TGRR260 pKa = 11.84LVLDD264 pKa = 3.95AAMDD268 pKa = 4.08PRR270 pKa = 11.84LSTLDD275 pKa = 3.35TDD277 pKa = 4.37RR278 pKa = 11.84EE279 pKa = 4.1QAGGFEE285 pKa = 4.07TAFRR289 pKa = 11.84SFAEE293 pKa = 4.38DD294 pKa = 3.26CTPRR298 pKa = 11.84DD299 pKa = 3.45SCALGNDD306 pKa = 4.17GADD309 pKa = 3.7GASQALLDD317 pKa = 4.49FFAEE321 pKa = 4.05VDD323 pKa = 3.92AEE325 pKa = 4.09PLPTGDD331 pKa = 3.71EE332 pKa = 4.09EE333 pKa = 5.36RR334 pKa = 11.84PLTEE338 pKa = 4.23SLASTGVSYY347 pKa = 11.45ALYY350 pKa = 10.8AEE352 pKa = 5.14DD353 pKa = 4.28LWPMLRR359 pKa = 11.84DD360 pKa = 3.64ALATAIEE367 pKa = 4.94DD368 pKa = 3.94GDD370 pKa = 4.13GSALLDD376 pKa = 4.1LADD379 pKa = 4.61AYY381 pKa = 11.05NEE383 pKa = 4.08RR384 pKa = 11.84EE385 pKa = 4.12ANGTYY390 pKa = 8.32GTTMFAFPAISCLDD404 pKa = 3.61SPAGNEE410 pKa = 3.87DD411 pKa = 3.31ADD413 pKa = 3.99AVRR416 pKa = 11.84SALAAYY422 pKa = 9.37EE423 pKa = 4.11EE424 pKa = 4.49ASPTFGRR431 pKa = 11.84DD432 pKa = 3.42FAWATLLCAAWPVDD446 pKa = 3.61PSGAPVSIPAADD458 pKa = 3.45ATDD461 pKa = 3.22ILVIGTLRR469 pKa = 11.84DD470 pKa = 3.29PATPYY475 pKa = 10.98AWAEE479 pKa = 3.9GLADD483 pKa = 3.68QLEE486 pKa = 4.49TGILLTYY493 pKa = 10.73DD494 pKa = 3.68GDD496 pKa = 3.74GHH498 pKa = 6.1GAYY501 pKa = 9.86GGASEE506 pKa = 5.15CVDD509 pKa = 3.56SAVNDD514 pKa = 3.69YY515 pKa = 11.17LLEE518 pKa = 4.39NVSPEE523 pKa = 4.96DD524 pKa = 3.5GTTCC528 pKa = 4.71
Molecular weight: 55.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V2RK93|A0A1V2RK93_9ACTN Phosphotransferase enzyme family protein OS=Streptomyces sp. MP131-18 OX=1857892 GN=STBA_36440 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.81GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.81GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2289126 |
29 |
4553 |
325.3 |
34.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.292 ± 0.054 | 0.799 ± 0.008 |
6.014 ± 0.024 | 5.936 ± 0.025 |
2.689 ± 0.016 | 9.761 ± 0.036 |
2.407 ± 0.016 | 3.022 ± 0.021 |
1.396 ± 0.016 | 10.43 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.683 ± 0.012 | 1.672 ± 0.016 |
6.369 ± 0.03 | 2.659 ± 0.02 |
8.638 ± 0.036 | 4.695 ± 0.025 |
5.883 ± 0.027 | 8.138 ± 0.028 |
1.569 ± 0.012 | 1.946 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
