
Cronartium ribicola mitovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
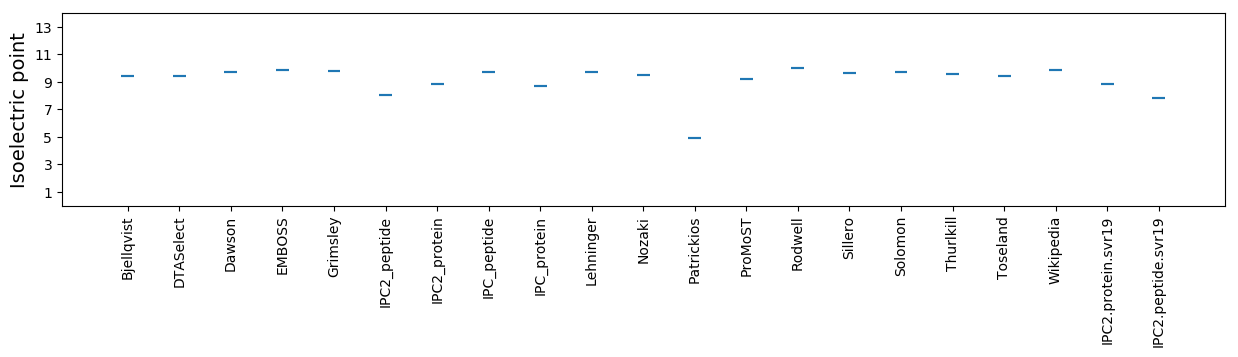
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191KCN6|A0A191KCN6_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 2 OX=1816485 GN=RdRp PE=4 SV=1
MM1 pKa = 7.66NIQAMMRR8 pKa = 11.84WLAVHH13 pKa = 6.54HH14 pKa = 6.4SCSPEE19 pKa = 3.63LMNVVEE25 pKa = 4.44TVLIKK30 pKa = 10.01WDD32 pKa = 3.66LLVRR36 pKa = 11.84TKK38 pKa = 10.83GIKK41 pKa = 8.79EE42 pKa = 3.97TVKK45 pKa = 10.28YY46 pKa = 9.66FKK48 pKa = 10.56ASRR51 pKa = 11.84LHH53 pKa = 5.01CTRR56 pKa = 11.84YY57 pKa = 9.74ICGDD61 pKa = 3.69PLLVSEE67 pKa = 5.87GISISKK73 pKa = 10.56DD74 pKa = 3.14GLPNLIKK81 pKa = 10.83GHH83 pKa = 5.79GKK85 pKa = 9.91SFFRR89 pKa = 11.84NKK91 pKa = 9.86KK92 pKa = 8.38INEE95 pKa = 3.45IRR97 pKa = 11.84IILTILSMGRR107 pKa = 11.84IIPGTGEE114 pKa = 4.13CDD116 pKa = 3.09LTPISKK122 pKa = 9.27PWLGQIPTSITRR134 pKa = 11.84WINSRR139 pKa = 11.84TDD141 pKa = 2.87ISKK144 pKa = 10.66VEE146 pKa = 4.09IEE148 pKa = 4.08WKK150 pKa = 9.46EE151 pKa = 3.89PHH153 pKa = 6.85WSTKK157 pKa = 10.46AGPKK161 pKa = 9.15GQAIASSIEE170 pKa = 4.02DD171 pKa = 3.66LASLPEE177 pKa = 3.98TLRR180 pKa = 11.84TDD182 pKa = 2.46IRR184 pKa = 11.84TIGGEE189 pKa = 4.06EE190 pKa = 3.75LSEE193 pKa = 4.39YY194 pKa = 10.34IDD196 pKa = 3.66TLEE199 pKa = 4.5PYY201 pKa = 10.08YY202 pKa = 11.12KK203 pKa = 10.66NILPNPKK210 pKa = 9.35RR211 pKa = 11.84RR212 pKa = 11.84SKK214 pKa = 10.34LASSIRR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 5.95IVLDD227 pKa = 3.81KK228 pKa = 10.6EE229 pKa = 4.49YY230 pKa = 8.72KK231 pKa = 8.37TRR233 pKa = 11.84PIAIIDD239 pKa = 3.63YY240 pKa = 8.68WSQTSLVPYY249 pKa = 9.62HH250 pKa = 7.23DD251 pKa = 5.73YY252 pKa = 10.44IMKK255 pKa = 9.58WIQKK259 pKa = 9.93QYY261 pKa = 11.54GDD263 pKa = 3.64YY264 pKa = 10.11TLQSKK269 pKa = 9.89VGKK272 pKa = 10.31QIVKK276 pKa = 9.15GQGRR280 pKa = 11.84IYY282 pKa = 11.01SFDD285 pKa = 3.6LQNATDD291 pKa = 4.16RR292 pKa = 11.84FPLEE296 pKa = 3.5FQKK299 pKa = 10.72IVFAKK304 pKa = 10.32FFGSKK309 pKa = 10.15KK310 pKa = 8.73ADD312 pKa = 2.63AWARR316 pKa = 11.84IITQYY321 pKa = 10.9PFEE324 pKa = 5.2LKK326 pKa = 10.61DD327 pKa = 3.67GSTVSYY333 pKa = 11.27NCGQPIGAYY342 pKa = 9.7SSWAIFTASHH352 pKa = 6.39HH353 pKa = 7.09LILQYY358 pKa = 10.55IRR360 pKa = 11.84FIHH363 pKa = 6.96PEE365 pKa = 3.93TIYY368 pKa = 11.0GVLGDD373 pKa = 4.35DD374 pKa = 3.6VVIRR378 pKa = 11.84GDD380 pKa = 3.3RR381 pKa = 11.84GAFLYY386 pKa = 10.63KK387 pKa = 10.44EE388 pKa = 4.36VMSCLDD394 pKa = 3.48VKK396 pKa = 10.74ISEE399 pKa = 4.65SKK401 pKa = 8.33THH403 pKa = 5.58VSNDD407 pKa = 2.86TFEE410 pKa = 4.1FAKK413 pKa = 10.35RR414 pKa = 11.84WFHH417 pKa = 6.74KK418 pKa = 10.35GAEE421 pKa = 4.13ITPFPLNEE429 pKa = 4.82AIEE432 pKa = 4.63AGTDD436 pKa = 3.35VAKK439 pKa = 10.8ASQLFLNLPLRR450 pKa = 11.84GWYY453 pKa = 10.29LGGNDD458 pKa = 3.99LSQSILEE465 pKa = 4.12FMKK468 pKa = 10.94SKK470 pKa = 10.33IVPSNFASYY479 pKa = 10.36LARR482 pKa = 11.84RR483 pKa = 11.84SARR486 pKa = 11.84FIYY489 pKa = 8.42FQKK492 pKa = 10.18WAITGCYY499 pKa = 9.74SYY501 pKa = 11.2LSKK504 pKa = 10.84LHH506 pKa = 6.37PLVLVSGSSVCNHH519 pKa = 5.5TEE521 pKa = 3.9RR522 pKa = 11.84LRR524 pKa = 11.84TAIMLGISKK533 pKa = 8.6ATAKK537 pKa = 10.31EE538 pKa = 3.65ISQILEE544 pKa = 4.81DD545 pKa = 4.5INTLLKK551 pKa = 10.73RR552 pKa = 11.84KK553 pKa = 9.73DD554 pKa = 3.24IQGLVDD560 pKa = 3.48QTTDD564 pKa = 3.11SPSYY568 pKa = 9.12TDD570 pKa = 4.2LRR572 pKa = 11.84PGEE575 pKa = 4.1YY576 pKa = 10.51ALGKK580 pKa = 10.26ISLKK584 pKa = 10.03LQKK587 pKa = 9.77EE588 pKa = 4.12LRR590 pKa = 11.84QFDD593 pKa = 3.78VDD595 pKa = 2.89IHH597 pKa = 6.03MLSKK601 pKa = 10.13MAAVPDD607 pKa = 3.97WATHH611 pKa = 5.7LGEE614 pKa = 4.78ASISKK619 pKa = 10.47LKK621 pKa = 10.33GIPVGRR627 pKa = 11.84RR628 pKa = 11.84PDD630 pKa = 3.76PDD632 pKa = 3.54TYY634 pKa = 10.23ISEE637 pKa = 4.17RR638 pKa = 11.84RR639 pKa = 11.84AVTATRR645 pKa = 11.84SLLRR649 pKa = 11.84VYY651 pKa = 11.35NNFNTEE657 pKa = 3.95TQQFFAIRR665 pKa = 11.84PAIHH669 pKa = 6.9RR670 pKa = 11.84PDD672 pKa = 3.51ANKK675 pKa = 10.61SIGSHH680 pKa = 4.96VRR682 pKa = 11.84TAAQAISS689 pKa = 3.26
MM1 pKa = 7.66NIQAMMRR8 pKa = 11.84WLAVHH13 pKa = 6.54HH14 pKa = 6.4SCSPEE19 pKa = 3.63LMNVVEE25 pKa = 4.44TVLIKK30 pKa = 10.01WDD32 pKa = 3.66LLVRR36 pKa = 11.84TKK38 pKa = 10.83GIKK41 pKa = 8.79EE42 pKa = 3.97TVKK45 pKa = 10.28YY46 pKa = 9.66FKK48 pKa = 10.56ASRR51 pKa = 11.84LHH53 pKa = 5.01CTRR56 pKa = 11.84YY57 pKa = 9.74ICGDD61 pKa = 3.69PLLVSEE67 pKa = 5.87GISISKK73 pKa = 10.56DD74 pKa = 3.14GLPNLIKK81 pKa = 10.83GHH83 pKa = 5.79GKK85 pKa = 9.91SFFRR89 pKa = 11.84NKK91 pKa = 9.86KK92 pKa = 8.38INEE95 pKa = 3.45IRR97 pKa = 11.84IILTILSMGRR107 pKa = 11.84IIPGTGEE114 pKa = 4.13CDD116 pKa = 3.09LTPISKK122 pKa = 9.27PWLGQIPTSITRR134 pKa = 11.84WINSRR139 pKa = 11.84TDD141 pKa = 2.87ISKK144 pKa = 10.66VEE146 pKa = 4.09IEE148 pKa = 4.08WKK150 pKa = 9.46EE151 pKa = 3.89PHH153 pKa = 6.85WSTKK157 pKa = 10.46AGPKK161 pKa = 9.15GQAIASSIEE170 pKa = 4.02DD171 pKa = 3.66LASLPEE177 pKa = 3.98TLRR180 pKa = 11.84TDD182 pKa = 2.46IRR184 pKa = 11.84TIGGEE189 pKa = 4.06EE190 pKa = 3.75LSEE193 pKa = 4.39YY194 pKa = 10.34IDD196 pKa = 3.66TLEE199 pKa = 4.5PYY201 pKa = 10.08YY202 pKa = 11.12KK203 pKa = 10.66NILPNPKK210 pKa = 9.35RR211 pKa = 11.84RR212 pKa = 11.84SKK214 pKa = 10.34LASSIRR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 5.95IVLDD227 pKa = 3.81KK228 pKa = 10.6EE229 pKa = 4.49YY230 pKa = 8.72KK231 pKa = 8.37TRR233 pKa = 11.84PIAIIDD239 pKa = 3.63YY240 pKa = 8.68WSQTSLVPYY249 pKa = 9.62HH250 pKa = 7.23DD251 pKa = 5.73YY252 pKa = 10.44IMKK255 pKa = 9.58WIQKK259 pKa = 9.93QYY261 pKa = 11.54GDD263 pKa = 3.64YY264 pKa = 10.11TLQSKK269 pKa = 9.89VGKK272 pKa = 10.31QIVKK276 pKa = 9.15GQGRR280 pKa = 11.84IYY282 pKa = 11.01SFDD285 pKa = 3.6LQNATDD291 pKa = 4.16RR292 pKa = 11.84FPLEE296 pKa = 3.5FQKK299 pKa = 10.72IVFAKK304 pKa = 10.32FFGSKK309 pKa = 10.15KK310 pKa = 8.73ADD312 pKa = 2.63AWARR316 pKa = 11.84IITQYY321 pKa = 10.9PFEE324 pKa = 5.2LKK326 pKa = 10.61DD327 pKa = 3.67GSTVSYY333 pKa = 11.27NCGQPIGAYY342 pKa = 9.7SSWAIFTASHH352 pKa = 6.39HH353 pKa = 7.09LILQYY358 pKa = 10.55IRR360 pKa = 11.84FIHH363 pKa = 6.96PEE365 pKa = 3.93TIYY368 pKa = 11.0GVLGDD373 pKa = 4.35DD374 pKa = 3.6VVIRR378 pKa = 11.84GDD380 pKa = 3.3RR381 pKa = 11.84GAFLYY386 pKa = 10.63KK387 pKa = 10.44EE388 pKa = 4.36VMSCLDD394 pKa = 3.48VKK396 pKa = 10.74ISEE399 pKa = 4.65SKK401 pKa = 8.33THH403 pKa = 5.58VSNDD407 pKa = 2.86TFEE410 pKa = 4.1FAKK413 pKa = 10.35RR414 pKa = 11.84WFHH417 pKa = 6.74KK418 pKa = 10.35GAEE421 pKa = 4.13ITPFPLNEE429 pKa = 4.82AIEE432 pKa = 4.63AGTDD436 pKa = 3.35VAKK439 pKa = 10.8ASQLFLNLPLRR450 pKa = 11.84GWYY453 pKa = 10.29LGGNDD458 pKa = 3.99LSQSILEE465 pKa = 4.12FMKK468 pKa = 10.94SKK470 pKa = 10.33IVPSNFASYY479 pKa = 10.36LARR482 pKa = 11.84RR483 pKa = 11.84SARR486 pKa = 11.84FIYY489 pKa = 8.42FQKK492 pKa = 10.18WAITGCYY499 pKa = 9.74SYY501 pKa = 11.2LSKK504 pKa = 10.84LHH506 pKa = 6.37PLVLVSGSSVCNHH519 pKa = 5.5TEE521 pKa = 3.9RR522 pKa = 11.84LRR524 pKa = 11.84TAIMLGISKK533 pKa = 8.6ATAKK537 pKa = 10.31EE538 pKa = 3.65ISQILEE544 pKa = 4.81DD545 pKa = 4.5INTLLKK551 pKa = 10.73RR552 pKa = 11.84KK553 pKa = 9.73DD554 pKa = 3.24IQGLVDD560 pKa = 3.48QTTDD564 pKa = 3.11SPSYY568 pKa = 9.12TDD570 pKa = 4.2LRR572 pKa = 11.84PGEE575 pKa = 4.1YY576 pKa = 10.51ALGKK580 pKa = 10.26ISLKK584 pKa = 10.03LQKK587 pKa = 9.77EE588 pKa = 4.12LRR590 pKa = 11.84QFDD593 pKa = 3.78VDD595 pKa = 2.89IHH597 pKa = 6.03MLSKK601 pKa = 10.13MAAVPDD607 pKa = 3.97WATHH611 pKa = 5.7LGEE614 pKa = 4.78ASISKK619 pKa = 10.47LKK621 pKa = 10.33GIPVGRR627 pKa = 11.84RR628 pKa = 11.84PDD630 pKa = 3.76PDD632 pKa = 3.54TYY634 pKa = 10.23ISEE637 pKa = 4.17RR638 pKa = 11.84RR639 pKa = 11.84AVTATRR645 pKa = 11.84SLLRR649 pKa = 11.84VYY651 pKa = 11.35NNFNTEE657 pKa = 3.95TQQFFAIRR665 pKa = 11.84PAIHH669 pKa = 6.9RR670 pKa = 11.84PDD672 pKa = 3.51ANKK675 pKa = 10.61SIGSHH680 pKa = 4.96VRR682 pKa = 11.84TAAQAISS689 pKa = 3.26
Molecular weight: 78.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191KCN6|A0A191KCN6_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 2 OX=1816485 GN=RdRp PE=4 SV=1
MM1 pKa = 7.66NIQAMMRR8 pKa = 11.84WLAVHH13 pKa = 6.54HH14 pKa = 6.4SCSPEE19 pKa = 3.63LMNVVEE25 pKa = 4.44TVLIKK30 pKa = 10.01WDD32 pKa = 3.66LLVRR36 pKa = 11.84TKK38 pKa = 10.83GIKK41 pKa = 8.79EE42 pKa = 3.97TVKK45 pKa = 10.28YY46 pKa = 9.66FKK48 pKa = 10.56ASRR51 pKa = 11.84LHH53 pKa = 5.01CTRR56 pKa = 11.84YY57 pKa = 9.74ICGDD61 pKa = 3.69PLLVSEE67 pKa = 5.87GISISKK73 pKa = 10.56DD74 pKa = 3.14GLPNLIKK81 pKa = 10.83GHH83 pKa = 5.79GKK85 pKa = 9.91SFFRR89 pKa = 11.84NKK91 pKa = 9.86KK92 pKa = 8.38INEE95 pKa = 3.45IRR97 pKa = 11.84IILTILSMGRR107 pKa = 11.84IIPGTGEE114 pKa = 4.13CDD116 pKa = 3.09LTPISKK122 pKa = 9.27PWLGQIPTSITRR134 pKa = 11.84WINSRR139 pKa = 11.84TDD141 pKa = 2.87ISKK144 pKa = 10.66VEE146 pKa = 4.09IEE148 pKa = 4.08WKK150 pKa = 9.46EE151 pKa = 3.89PHH153 pKa = 6.85WSTKK157 pKa = 10.46AGPKK161 pKa = 9.15GQAIASSIEE170 pKa = 4.02DD171 pKa = 3.66LASLPEE177 pKa = 3.98TLRR180 pKa = 11.84TDD182 pKa = 2.46IRR184 pKa = 11.84TIGGEE189 pKa = 4.06EE190 pKa = 3.75LSEE193 pKa = 4.39YY194 pKa = 10.34IDD196 pKa = 3.66TLEE199 pKa = 4.5PYY201 pKa = 10.08YY202 pKa = 11.12KK203 pKa = 10.66NILPNPKK210 pKa = 9.35RR211 pKa = 11.84RR212 pKa = 11.84SKK214 pKa = 10.34LASSIRR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 5.95IVLDD227 pKa = 3.81KK228 pKa = 10.6EE229 pKa = 4.49YY230 pKa = 8.72KK231 pKa = 8.37TRR233 pKa = 11.84PIAIIDD239 pKa = 3.63YY240 pKa = 8.68WSQTSLVPYY249 pKa = 9.62HH250 pKa = 7.23DD251 pKa = 5.73YY252 pKa = 10.44IMKK255 pKa = 9.58WIQKK259 pKa = 9.93QYY261 pKa = 11.54GDD263 pKa = 3.64YY264 pKa = 10.11TLQSKK269 pKa = 9.89VGKK272 pKa = 10.31QIVKK276 pKa = 9.15GQGRR280 pKa = 11.84IYY282 pKa = 11.01SFDD285 pKa = 3.6LQNATDD291 pKa = 4.16RR292 pKa = 11.84FPLEE296 pKa = 3.5FQKK299 pKa = 10.72IVFAKK304 pKa = 10.32FFGSKK309 pKa = 10.15KK310 pKa = 8.73ADD312 pKa = 2.63AWARR316 pKa = 11.84IITQYY321 pKa = 10.9PFEE324 pKa = 5.2LKK326 pKa = 10.61DD327 pKa = 3.67GSTVSYY333 pKa = 11.27NCGQPIGAYY342 pKa = 9.7SSWAIFTASHH352 pKa = 6.39HH353 pKa = 7.09LILQYY358 pKa = 10.55IRR360 pKa = 11.84FIHH363 pKa = 6.96PEE365 pKa = 3.93TIYY368 pKa = 11.0GVLGDD373 pKa = 4.35DD374 pKa = 3.6VVIRR378 pKa = 11.84GDD380 pKa = 3.3RR381 pKa = 11.84GAFLYY386 pKa = 10.63KK387 pKa = 10.44EE388 pKa = 4.36VMSCLDD394 pKa = 3.48VKK396 pKa = 10.74ISEE399 pKa = 4.65SKK401 pKa = 8.33THH403 pKa = 5.58VSNDD407 pKa = 2.86TFEE410 pKa = 4.1FAKK413 pKa = 10.35RR414 pKa = 11.84WFHH417 pKa = 6.74KK418 pKa = 10.35GAEE421 pKa = 4.13ITPFPLNEE429 pKa = 4.82AIEE432 pKa = 4.63AGTDD436 pKa = 3.35VAKK439 pKa = 10.8ASQLFLNLPLRR450 pKa = 11.84GWYY453 pKa = 10.29LGGNDD458 pKa = 3.99LSQSILEE465 pKa = 4.12FMKK468 pKa = 10.94SKK470 pKa = 10.33IVPSNFASYY479 pKa = 10.36LARR482 pKa = 11.84RR483 pKa = 11.84SARR486 pKa = 11.84FIYY489 pKa = 8.42FQKK492 pKa = 10.18WAITGCYY499 pKa = 9.74SYY501 pKa = 11.2LSKK504 pKa = 10.84LHH506 pKa = 6.37PLVLVSGSSVCNHH519 pKa = 5.5TEE521 pKa = 3.9RR522 pKa = 11.84LRR524 pKa = 11.84TAIMLGISKK533 pKa = 8.6ATAKK537 pKa = 10.31EE538 pKa = 3.65ISQILEE544 pKa = 4.81DD545 pKa = 4.5INTLLKK551 pKa = 10.73RR552 pKa = 11.84KK553 pKa = 9.73DD554 pKa = 3.24IQGLVDD560 pKa = 3.48QTTDD564 pKa = 3.11SPSYY568 pKa = 9.12TDD570 pKa = 4.2LRR572 pKa = 11.84PGEE575 pKa = 4.1YY576 pKa = 10.51ALGKK580 pKa = 10.26ISLKK584 pKa = 10.03LQKK587 pKa = 9.77EE588 pKa = 4.12LRR590 pKa = 11.84QFDD593 pKa = 3.78VDD595 pKa = 2.89IHH597 pKa = 6.03MLSKK601 pKa = 10.13MAAVPDD607 pKa = 3.97WATHH611 pKa = 5.7LGEE614 pKa = 4.78ASISKK619 pKa = 10.47LKK621 pKa = 10.33GIPVGRR627 pKa = 11.84RR628 pKa = 11.84PDD630 pKa = 3.76PDD632 pKa = 3.54TYY634 pKa = 10.23ISEE637 pKa = 4.17RR638 pKa = 11.84RR639 pKa = 11.84AVTATRR645 pKa = 11.84SLLRR649 pKa = 11.84VYY651 pKa = 11.35NNFNTEE657 pKa = 3.95TQQFFAIRR665 pKa = 11.84PAIHH669 pKa = 6.9RR670 pKa = 11.84PDD672 pKa = 3.51ANKK675 pKa = 10.61SIGSHH680 pKa = 4.96VRR682 pKa = 11.84TAAQAISS689 pKa = 3.26
MM1 pKa = 7.66NIQAMMRR8 pKa = 11.84WLAVHH13 pKa = 6.54HH14 pKa = 6.4SCSPEE19 pKa = 3.63LMNVVEE25 pKa = 4.44TVLIKK30 pKa = 10.01WDD32 pKa = 3.66LLVRR36 pKa = 11.84TKK38 pKa = 10.83GIKK41 pKa = 8.79EE42 pKa = 3.97TVKK45 pKa = 10.28YY46 pKa = 9.66FKK48 pKa = 10.56ASRR51 pKa = 11.84LHH53 pKa = 5.01CTRR56 pKa = 11.84YY57 pKa = 9.74ICGDD61 pKa = 3.69PLLVSEE67 pKa = 5.87GISISKK73 pKa = 10.56DD74 pKa = 3.14GLPNLIKK81 pKa = 10.83GHH83 pKa = 5.79GKK85 pKa = 9.91SFFRR89 pKa = 11.84NKK91 pKa = 9.86KK92 pKa = 8.38INEE95 pKa = 3.45IRR97 pKa = 11.84IILTILSMGRR107 pKa = 11.84IIPGTGEE114 pKa = 4.13CDD116 pKa = 3.09LTPISKK122 pKa = 9.27PWLGQIPTSITRR134 pKa = 11.84WINSRR139 pKa = 11.84TDD141 pKa = 2.87ISKK144 pKa = 10.66VEE146 pKa = 4.09IEE148 pKa = 4.08WKK150 pKa = 9.46EE151 pKa = 3.89PHH153 pKa = 6.85WSTKK157 pKa = 10.46AGPKK161 pKa = 9.15GQAIASSIEE170 pKa = 4.02DD171 pKa = 3.66LASLPEE177 pKa = 3.98TLRR180 pKa = 11.84TDD182 pKa = 2.46IRR184 pKa = 11.84TIGGEE189 pKa = 4.06EE190 pKa = 3.75LSEE193 pKa = 4.39YY194 pKa = 10.34IDD196 pKa = 3.66TLEE199 pKa = 4.5PYY201 pKa = 10.08YY202 pKa = 11.12KK203 pKa = 10.66NILPNPKK210 pKa = 9.35RR211 pKa = 11.84RR212 pKa = 11.84SKK214 pKa = 10.34LASSIRR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 5.95IVLDD227 pKa = 3.81KK228 pKa = 10.6EE229 pKa = 4.49YY230 pKa = 8.72KK231 pKa = 8.37TRR233 pKa = 11.84PIAIIDD239 pKa = 3.63YY240 pKa = 8.68WSQTSLVPYY249 pKa = 9.62HH250 pKa = 7.23DD251 pKa = 5.73YY252 pKa = 10.44IMKK255 pKa = 9.58WIQKK259 pKa = 9.93QYY261 pKa = 11.54GDD263 pKa = 3.64YY264 pKa = 10.11TLQSKK269 pKa = 9.89VGKK272 pKa = 10.31QIVKK276 pKa = 9.15GQGRR280 pKa = 11.84IYY282 pKa = 11.01SFDD285 pKa = 3.6LQNATDD291 pKa = 4.16RR292 pKa = 11.84FPLEE296 pKa = 3.5FQKK299 pKa = 10.72IVFAKK304 pKa = 10.32FFGSKK309 pKa = 10.15KK310 pKa = 8.73ADD312 pKa = 2.63AWARR316 pKa = 11.84IITQYY321 pKa = 10.9PFEE324 pKa = 5.2LKK326 pKa = 10.61DD327 pKa = 3.67GSTVSYY333 pKa = 11.27NCGQPIGAYY342 pKa = 9.7SSWAIFTASHH352 pKa = 6.39HH353 pKa = 7.09LILQYY358 pKa = 10.55IRR360 pKa = 11.84FIHH363 pKa = 6.96PEE365 pKa = 3.93TIYY368 pKa = 11.0GVLGDD373 pKa = 4.35DD374 pKa = 3.6VVIRR378 pKa = 11.84GDD380 pKa = 3.3RR381 pKa = 11.84GAFLYY386 pKa = 10.63KK387 pKa = 10.44EE388 pKa = 4.36VMSCLDD394 pKa = 3.48VKK396 pKa = 10.74ISEE399 pKa = 4.65SKK401 pKa = 8.33THH403 pKa = 5.58VSNDD407 pKa = 2.86TFEE410 pKa = 4.1FAKK413 pKa = 10.35RR414 pKa = 11.84WFHH417 pKa = 6.74KK418 pKa = 10.35GAEE421 pKa = 4.13ITPFPLNEE429 pKa = 4.82AIEE432 pKa = 4.63AGTDD436 pKa = 3.35VAKK439 pKa = 10.8ASQLFLNLPLRR450 pKa = 11.84GWYY453 pKa = 10.29LGGNDD458 pKa = 3.99LSQSILEE465 pKa = 4.12FMKK468 pKa = 10.94SKK470 pKa = 10.33IVPSNFASYY479 pKa = 10.36LARR482 pKa = 11.84RR483 pKa = 11.84SARR486 pKa = 11.84FIYY489 pKa = 8.42FQKK492 pKa = 10.18WAITGCYY499 pKa = 9.74SYY501 pKa = 11.2LSKK504 pKa = 10.84LHH506 pKa = 6.37PLVLVSGSSVCNHH519 pKa = 5.5TEE521 pKa = 3.9RR522 pKa = 11.84LRR524 pKa = 11.84TAIMLGISKK533 pKa = 8.6ATAKK537 pKa = 10.31EE538 pKa = 3.65ISQILEE544 pKa = 4.81DD545 pKa = 4.5INTLLKK551 pKa = 10.73RR552 pKa = 11.84KK553 pKa = 9.73DD554 pKa = 3.24IQGLVDD560 pKa = 3.48QTTDD564 pKa = 3.11SPSYY568 pKa = 9.12TDD570 pKa = 4.2LRR572 pKa = 11.84PGEE575 pKa = 4.1YY576 pKa = 10.51ALGKK580 pKa = 10.26ISLKK584 pKa = 10.03LQKK587 pKa = 9.77EE588 pKa = 4.12LRR590 pKa = 11.84QFDD593 pKa = 3.78VDD595 pKa = 2.89IHH597 pKa = 6.03MLSKK601 pKa = 10.13MAAVPDD607 pKa = 3.97WATHH611 pKa = 5.7LGEE614 pKa = 4.78ASISKK619 pKa = 10.47LKK621 pKa = 10.33GIPVGRR627 pKa = 11.84RR628 pKa = 11.84PDD630 pKa = 3.76PDD632 pKa = 3.54TYY634 pKa = 10.23ISEE637 pKa = 4.17RR638 pKa = 11.84RR639 pKa = 11.84AVTATRR645 pKa = 11.84SLLRR649 pKa = 11.84VYY651 pKa = 11.35NNFNTEE657 pKa = 3.95TQQFFAIRR665 pKa = 11.84PAIHH669 pKa = 6.9RR670 pKa = 11.84PDD672 pKa = 3.51ANKK675 pKa = 10.61SIGSHH680 pKa = 4.96VRR682 pKa = 11.84TAAQAISS689 pKa = 3.26
Molecular weight: 78.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
689 |
689 |
689 |
689.0 |
78.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.386 ± 0.0 | 1.161 ± 0.0 |
4.935 ± 0.0 | 4.935 ± 0.0 |
3.774 ± 0.0 | 5.951 ± 0.0 |
2.612 ± 0.0 | 9.724 ± 0.0 |
7.402 ± 0.0 | 8.999 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.597 ± 0.0 | 3.048 ± 0.0 |
4.644 ± 0.0 | 3.628 ± 0.0 |
5.951 ± 0.0 | 8.563 ± 0.0 |
6.096 ± 0.0 | 4.644 ± 0.0 |
2.032 ± 0.0 | 3.919 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
