
Turkey associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
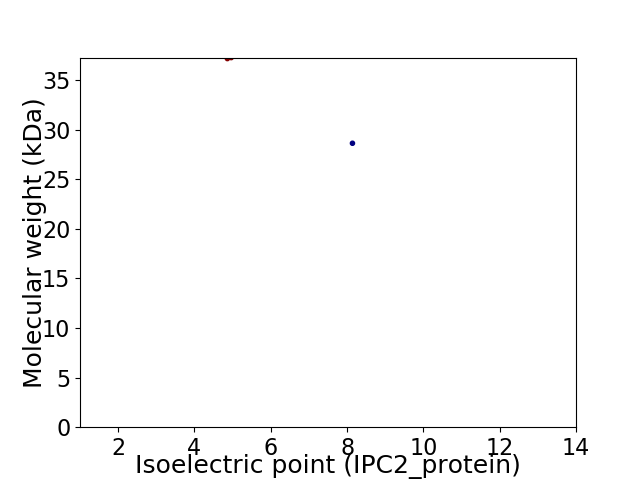
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0GEQ0|W0GEQ0_9VIRU Capsid protein OS=Turkey associated porprismacovirus 1 OX=2170130 PE=4 SV=1
MM1 pKa = 6.48VAVRR5 pKa = 11.84VSEE8 pKa = 4.31TYY10 pKa = 10.76DD11 pKa = 3.27LSTKK15 pKa = 9.04IGKK18 pKa = 8.47MGLVGIHH25 pKa = 6.31TPTGDD30 pKa = 4.17LISRR34 pKa = 11.84MWPGLILQHH43 pKa = 6.04EE44 pKa = 4.62KK45 pKa = 10.79FRR47 pKa = 11.84FVKK50 pKa = 10.39CDD52 pKa = 2.95VAMACASMLPADD64 pKa = 4.26PLQIGVEE71 pKa = 4.06AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.62KK87 pKa = 10.55AVSNEE92 pKa = 4.08SMNQFQAWLNIVNQTAGLTGAFNQGSVVDD121 pKa = 4.11VNDD124 pKa = 4.13PNFVGANGTHH134 pKa = 6.93EE135 pKa = 4.16YY136 pKa = 10.78DD137 pKa = 3.44QFSMYY142 pKa = 10.32YY143 pKa = 10.72ALLADD148 pKa = 3.43SDD150 pKa = 4.11GWRR153 pKa = 11.84KK154 pKa = 10.33SMPQAGLEE162 pKa = 4.0MRR164 pKa = 11.84GLYY167 pKa = 9.85PLVFQVVSQFGSGPEE182 pKa = 3.96NSAGDD187 pKa = 3.93ADD189 pKa = 4.66SNISVPGLSGEE200 pKa = 4.37AADD203 pKa = 4.88ASPTSRR209 pKa = 11.84FGTVSSIQNRR219 pKa = 11.84TMRR222 pKa = 11.84GPSMRR227 pKa = 11.84MPAIDD232 pKa = 3.04TTFFVNSTSTDD243 pKa = 3.29SALLPGGINVDD254 pKa = 3.38TSLDD258 pKa = 3.94KK259 pKa = 11.4VPPAYY264 pKa = 10.27VGLIILPPARR274 pKa = 11.84LNQLYY279 pKa = 10.21YY280 pKa = 10.31RR281 pKa = 11.84LKK283 pKa = 8.78VTWTIEE289 pKa = 4.03FTGLRR294 pKa = 11.84SAVDD298 pKa = 2.97ISNWFNLQNIGQLAYY313 pKa = 10.38GSDD316 pKa = 4.08YY317 pKa = 10.93EE318 pKa = 4.37SQSAKK323 pKa = 9.97MSAKK327 pKa = 9.7TGMVDD332 pKa = 3.29TTGSSITKK340 pKa = 9.95IMEE343 pKa = 4.19GSS345 pKa = 3.18
MM1 pKa = 6.48VAVRR5 pKa = 11.84VSEE8 pKa = 4.31TYY10 pKa = 10.76DD11 pKa = 3.27LSTKK15 pKa = 9.04IGKK18 pKa = 8.47MGLVGIHH25 pKa = 6.31TPTGDD30 pKa = 4.17LISRR34 pKa = 11.84MWPGLILQHH43 pKa = 6.04EE44 pKa = 4.62KK45 pKa = 10.79FRR47 pKa = 11.84FVKK50 pKa = 10.39CDD52 pKa = 2.95VAMACASMLPADD64 pKa = 4.26PLQIGVEE71 pKa = 4.06AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.62KK87 pKa = 10.55AVSNEE92 pKa = 4.08SMNQFQAWLNIVNQTAGLTGAFNQGSVVDD121 pKa = 4.11VNDD124 pKa = 4.13PNFVGANGTHH134 pKa = 6.93EE135 pKa = 4.16YY136 pKa = 10.78DD137 pKa = 3.44QFSMYY142 pKa = 10.32YY143 pKa = 10.72ALLADD148 pKa = 3.43SDD150 pKa = 4.11GWRR153 pKa = 11.84KK154 pKa = 10.33SMPQAGLEE162 pKa = 4.0MRR164 pKa = 11.84GLYY167 pKa = 9.85PLVFQVVSQFGSGPEE182 pKa = 3.96NSAGDD187 pKa = 3.93ADD189 pKa = 4.66SNISVPGLSGEE200 pKa = 4.37AADD203 pKa = 4.88ASPTSRR209 pKa = 11.84FGTVSSIQNRR219 pKa = 11.84TMRR222 pKa = 11.84GPSMRR227 pKa = 11.84MPAIDD232 pKa = 3.04TTFFVNSTSTDD243 pKa = 3.29SALLPGGINVDD254 pKa = 3.38TSLDD258 pKa = 3.94KK259 pKa = 11.4VPPAYY264 pKa = 10.27VGLIILPPARR274 pKa = 11.84LNQLYY279 pKa = 10.21YY280 pKa = 10.31RR281 pKa = 11.84LKK283 pKa = 8.78VTWTIEE289 pKa = 4.03FTGLRR294 pKa = 11.84SAVDD298 pKa = 2.97ISNWFNLQNIGQLAYY313 pKa = 10.38GSDD316 pKa = 4.08YY317 pKa = 10.93EE318 pKa = 4.37SQSAKK323 pKa = 9.97MSAKK327 pKa = 9.7TGMVDD332 pKa = 3.29TTGSSITKK340 pKa = 9.95IMEE343 pKa = 4.19GSS345 pKa = 3.18
Molecular weight: 37.18 kDa
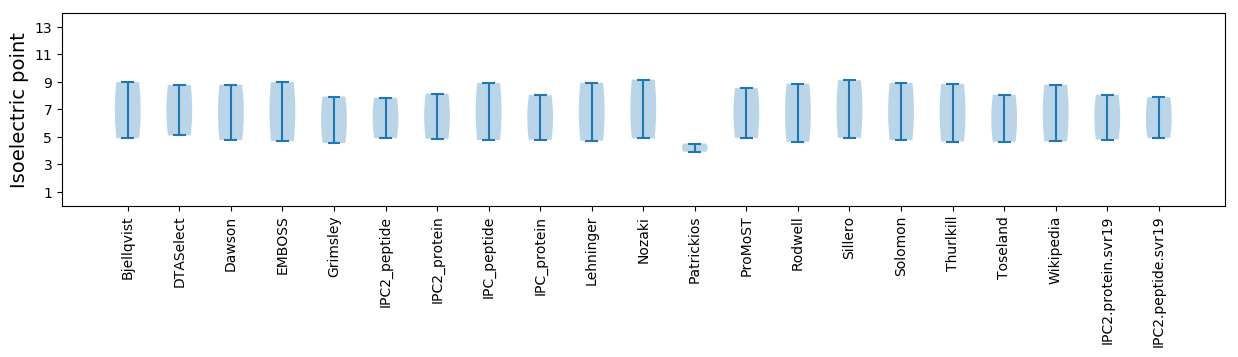
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0GEQ0|W0GEQ0_9VIRU Capsid protein OS=Turkey associated porprismacovirus 1 OX=2170130 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 9.88KK3 pKa = 10.66YY4 pKa = 10.74IMTVPRR10 pKa = 11.84SVPKK14 pKa = 10.24KK15 pKa = 9.7ALKK18 pKa = 10.51IMIDD22 pKa = 3.62VNDD25 pKa = 3.89CKK27 pKa = 10.84KK28 pKa = 10.09WIIGKK33 pKa = 8.89EE34 pKa = 3.81RR35 pKa = 11.84GKK37 pKa = 10.39NGYY40 pKa = 7.35EE41 pKa = 3.32HH42 pKa = 5.58WQIRR46 pKa = 11.84IEE48 pKa = 4.1TSNDD52 pKa = 3.12EE53 pKa = 3.95FFKK56 pKa = 10.0WCKK59 pKa = 9.35HH60 pKa = 6.21HH61 pKa = 7.13IPAAHH66 pKa = 6.86IEE68 pKa = 4.03EE69 pKa = 4.45AQQGVDD75 pKa = 2.8EE76 pKa = 4.66CLYY79 pKa = 10.31EE80 pKa = 4.45RR81 pKa = 11.84KK82 pKa = 9.86EE83 pKa = 4.17GQFWTSSDD91 pKa = 3.32RR92 pKa = 11.84VEE94 pKa = 4.28TLHH97 pKa = 6.68QRR99 pKa = 11.84FGTLRR104 pKa = 11.84PRR106 pKa = 11.84QEE108 pKa = 3.89RR109 pKa = 11.84ALLALQSTNDD119 pKa = 3.5RR120 pKa = 11.84EE121 pKa = 4.33VMVWYY126 pKa = 9.52DD127 pKa = 3.23ANGNVGKK134 pKa = 10.23SWFCGALWEE143 pKa = 4.8RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTVDD155 pKa = 3.07TVKK158 pKa = 11.13GLIQWVASCYY168 pKa = 9.46MDD170 pKa = 4.24GGWRR174 pKa = 11.84PYY176 pKa = 10.86VIIDD180 pKa = 4.59IPRR183 pKa = 11.84SWKK186 pKa = 9.98WSDD189 pKa = 3.42QLYY192 pKa = 10.52CAIEE196 pKa = 4.31SIKK199 pKa = 11.03DD200 pKa = 3.19GLIYY204 pKa = 9.12DD205 pKa = 4.11TRR207 pKa = 11.84YY208 pKa = 7.89HH209 pKa = 6.86ARR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 10.04VLVLTNTLPKK229 pKa = 9.94LDD231 pKa = 3.78KK232 pKa = 10.63LSRR235 pKa = 11.84DD236 pKa = 3.02RR237 pKa = 11.84WCIFEE242 pKa = 3.94FF243 pKa = 4.66
MM1 pKa = 7.58KK2 pKa = 9.88KK3 pKa = 10.66YY4 pKa = 10.74IMTVPRR10 pKa = 11.84SVPKK14 pKa = 10.24KK15 pKa = 9.7ALKK18 pKa = 10.51IMIDD22 pKa = 3.62VNDD25 pKa = 3.89CKK27 pKa = 10.84KK28 pKa = 10.09WIIGKK33 pKa = 8.89EE34 pKa = 3.81RR35 pKa = 11.84GKK37 pKa = 10.39NGYY40 pKa = 7.35EE41 pKa = 3.32HH42 pKa = 5.58WQIRR46 pKa = 11.84IEE48 pKa = 4.1TSNDD52 pKa = 3.12EE53 pKa = 3.95FFKK56 pKa = 10.0WCKK59 pKa = 9.35HH60 pKa = 6.21HH61 pKa = 7.13IPAAHH66 pKa = 6.86IEE68 pKa = 4.03EE69 pKa = 4.45AQQGVDD75 pKa = 2.8EE76 pKa = 4.66CLYY79 pKa = 10.31EE80 pKa = 4.45RR81 pKa = 11.84KK82 pKa = 9.86EE83 pKa = 4.17GQFWTSSDD91 pKa = 3.32RR92 pKa = 11.84VEE94 pKa = 4.28TLHH97 pKa = 6.68QRR99 pKa = 11.84FGTLRR104 pKa = 11.84PRR106 pKa = 11.84QEE108 pKa = 3.89RR109 pKa = 11.84ALLALQSTNDD119 pKa = 3.5RR120 pKa = 11.84EE121 pKa = 4.33VMVWYY126 pKa = 9.52DD127 pKa = 3.23ANGNVGKK134 pKa = 10.23SWFCGALWEE143 pKa = 4.8RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTVDD155 pKa = 3.07TVKK158 pKa = 11.13GLIQWVASCYY168 pKa = 9.46MDD170 pKa = 4.24GGWRR174 pKa = 11.84PYY176 pKa = 10.86VIIDD180 pKa = 4.59IPRR183 pKa = 11.84SWKK186 pKa = 9.98WSDD189 pKa = 3.42QLYY192 pKa = 10.52CAIEE196 pKa = 4.31SIKK199 pKa = 11.03DD200 pKa = 3.19GLIYY204 pKa = 9.12DD205 pKa = 4.11TRR207 pKa = 11.84YY208 pKa = 7.89HH209 pKa = 6.86ARR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 10.04VLVLTNTLPKK229 pKa = 9.94LDD231 pKa = 3.78KK232 pKa = 10.63LSRR235 pKa = 11.84DD236 pKa = 3.02RR237 pKa = 11.84WCIFEE242 pKa = 3.94FF243 pKa = 4.66
Molecular weight: 28.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
588 |
243 |
345 |
294.0 |
32.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.973 ± 1.164 | 1.531 ± 0.773 |
5.952 ± 0.126 | 4.422 ± 1.002 |
3.571 ± 0.395 | 7.823 ± 0.944 |
1.531 ± 0.537 | 6.463 ± 0.776 |
5.102 ± 1.555 | 7.483 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.741 ± 0.728 | 4.422 ± 0.646 |
4.762 ± 0.606 | 4.252 ± 0.314 |
5.102 ± 1.319 | 7.653 ± 1.789 |
5.782 ± 0.483 | 6.973 ± 0.222 |
2.891 ± 1.171 | 3.571 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
