
Brachybacterium phenoliresistens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermabacteraceae; Brachybacterium
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
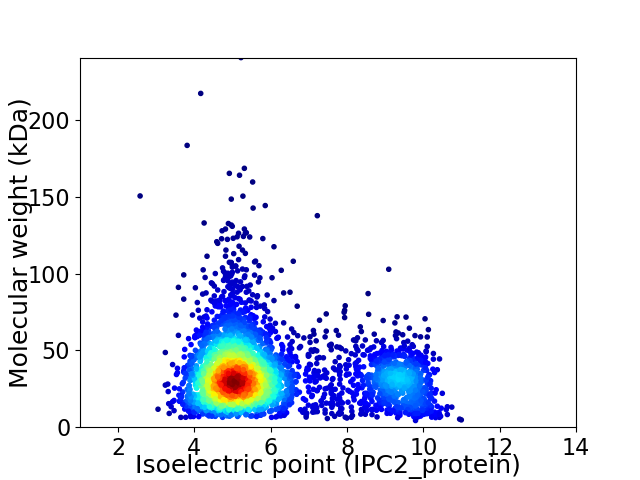
Virtual 2D-PAGE plot for 3605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Z9JRZ1|Z9JRZ1_9MICO RNA polymerase subunit sigma-24 OS=Brachybacterium phenoliresistens OX=396014 GN=BF93_02760 PE=3 SV=1
MM1 pKa = 7.6HH2 pKa = 7.81DD3 pKa = 3.61VRR5 pKa = 11.84HH6 pKa = 5.59GRR8 pKa = 11.84APRR11 pKa = 11.84AMRR14 pKa = 11.84YY15 pKa = 8.77LATAAAVLLVVAGCGPLDD33 pKa = 4.3RR34 pKa = 11.84IGIGGGGDD42 pKa = 3.61EE43 pKa = 4.49QTTAPAAGDD52 pKa = 3.5GTNAEE57 pKa = 4.63GEE59 pKa = 4.38ATPPLAEE66 pKa = 4.55TGTSAAAEE74 pKa = 4.32LPQDD78 pKa = 3.81PAGDD82 pKa = 3.56AAYY85 pKa = 10.0AAYY88 pKa = 10.49YY89 pKa = 8.75SQQVAWGPCDD99 pKa = 4.55DD100 pKa = 5.0SVDD103 pKa = 3.84GPASLEE109 pKa = 4.32CGTVTVPKK117 pKa = 10.25VWNDD121 pKa = 3.18PSAGDD126 pKa = 3.53LQIAVTRR133 pKa = 11.84LPATGQKK140 pKa = 9.8QGSLLTNPGGPGGSGVDD157 pKa = 4.41FVAGSAQTLFSEE169 pKa = 5.05EE170 pKa = 3.38VRR172 pKa = 11.84GAYY175 pKa = 10.08DD176 pKa = 3.05IVGFDD181 pKa = 3.43PRR183 pKa = 11.84GVSRR187 pKa = 11.84SAGITCLDD195 pKa = 3.89DD196 pKa = 5.28AEE198 pKa = 4.33TDD200 pKa = 3.17AYY202 pKa = 10.63RR203 pKa = 11.84AEE205 pKa = 4.39TFDD208 pKa = 4.13LADD211 pKa = 3.96PEE213 pKa = 4.4GLQAAEE219 pKa = 4.27DD220 pKa = 3.96AFTEE224 pKa = 5.0LSAACEE230 pKa = 4.16KK231 pKa = 11.05NSGDD235 pKa = 4.05LLPYY239 pKa = 10.36LDD241 pKa = 4.56TYY243 pKa = 11.16SAARR247 pKa = 11.84DD248 pKa = 3.52LDD250 pKa = 3.8VLRR253 pKa = 11.84SAVGSEE259 pKa = 3.45KK260 pKa = 10.42LDD262 pKa = 3.63YY263 pKa = 10.95VGYY266 pKa = 10.6SYY268 pKa = 10.44GTYY271 pKa = 10.47LGATYY276 pKa = 11.16AEE278 pKa = 4.83MYY280 pKa = 9.81PEE282 pKa = 3.79RR283 pKa = 11.84TGRR286 pKa = 11.84FVLDD290 pKa = 3.6GALDD294 pKa = 3.78PRR296 pKa = 11.84LTADD300 pKa = 5.23DD301 pKa = 3.58ITRR304 pKa = 11.84GQAEE308 pKa = 4.46GFEE311 pKa = 4.5EE312 pKa = 4.12ATSAFVEE319 pKa = 4.66SCLSQGSDD327 pKa = 3.31GCPLTGSVEE336 pKa = 4.17DD337 pKa = 4.0GKK339 pKa = 10.85EE340 pKa = 3.62QLRR343 pKa = 11.84AFFDD347 pKa = 3.86SVDD350 pKa = 3.7AQPLQTDD357 pKa = 3.8DD358 pKa = 4.04PNRR361 pKa = 11.84PLTGALARR369 pKa = 11.84SGLLVVLYY377 pKa = 10.7NDD379 pKa = 3.96EE380 pKa = 4.09NWPLGVQALNAAMTGDD396 pKa = 3.57GSTLLFLADD405 pKa = 4.21LSADD409 pKa = 3.96RR410 pKa = 11.84EE411 pKa = 4.24DD412 pKa = 4.4DD413 pKa = 3.33GTYY416 pKa = 10.54SGNGTFAITAVNCLDD431 pKa = 3.64HH432 pKa = 8.2VGVADD437 pKa = 3.92QQWQIDD443 pKa = 3.77EE444 pKa = 4.3AAAVAEE450 pKa = 4.8EE451 pKa = 4.45FPTWGPGFGFSQTMCDD467 pKa = 3.5LWPAKK472 pKa = 9.42PVRR475 pKa = 11.84EE476 pKa = 4.08PAPISAAGSDD486 pKa = 4.28LIVVIGTRR494 pKa = 11.84GDD496 pKa = 3.38PATPYY501 pKa = 10.55DD502 pKa = 3.56WAVGLDD508 pKa = 3.6EE509 pKa = 4.28QLEE512 pKa = 4.22NSTLITYY519 pKa = 9.63EE520 pKa = 4.34GEE522 pKa = 3.66GHH524 pKa = 5.38TAYY527 pKa = 10.17GRR529 pKa = 11.84SGGCVEE535 pKa = 4.59AAVDD539 pKa = 4.26AYY541 pKa = 10.68LLSGTAPEE549 pKa = 4.62EE550 pKa = 4.25GLTCSS555 pKa = 3.97
MM1 pKa = 7.6HH2 pKa = 7.81DD3 pKa = 3.61VRR5 pKa = 11.84HH6 pKa = 5.59GRR8 pKa = 11.84APRR11 pKa = 11.84AMRR14 pKa = 11.84YY15 pKa = 8.77LATAAAVLLVVAGCGPLDD33 pKa = 4.3RR34 pKa = 11.84IGIGGGGDD42 pKa = 3.61EE43 pKa = 4.49QTTAPAAGDD52 pKa = 3.5GTNAEE57 pKa = 4.63GEE59 pKa = 4.38ATPPLAEE66 pKa = 4.55TGTSAAAEE74 pKa = 4.32LPQDD78 pKa = 3.81PAGDD82 pKa = 3.56AAYY85 pKa = 10.0AAYY88 pKa = 10.49YY89 pKa = 8.75SQQVAWGPCDD99 pKa = 4.55DD100 pKa = 5.0SVDD103 pKa = 3.84GPASLEE109 pKa = 4.32CGTVTVPKK117 pKa = 10.25VWNDD121 pKa = 3.18PSAGDD126 pKa = 3.53LQIAVTRR133 pKa = 11.84LPATGQKK140 pKa = 9.8QGSLLTNPGGPGGSGVDD157 pKa = 4.41FVAGSAQTLFSEE169 pKa = 5.05EE170 pKa = 3.38VRR172 pKa = 11.84GAYY175 pKa = 10.08DD176 pKa = 3.05IVGFDD181 pKa = 3.43PRR183 pKa = 11.84GVSRR187 pKa = 11.84SAGITCLDD195 pKa = 3.89DD196 pKa = 5.28AEE198 pKa = 4.33TDD200 pKa = 3.17AYY202 pKa = 10.63RR203 pKa = 11.84AEE205 pKa = 4.39TFDD208 pKa = 4.13LADD211 pKa = 3.96PEE213 pKa = 4.4GLQAAEE219 pKa = 4.27DD220 pKa = 3.96AFTEE224 pKa = 5.0LSAACEE230 pKa = 4.16KK231 pKa = 11.05NSGDD235 pKa = 4.05LLPYY239 pKa = 10.36LDD241 pKa = 4.56TYY243 pKa = 11.16SAARR247 pKa = 11.84DD248 pKa = 3.52LDD250 pKa = 3.8VLRR253 pKa = 11.84SAVGSEE259 pKa = 3.45KK260 pKa = 10.42LDD262 pKa = 3.63YY263 pKa = 10.95VGYY266 pKa = 10.6SYY268 pKa = 10.44GTYY271 pKa = 10.47LGATYY276 pKa = 11.16AEE278 pKa = 4.83MYY280 pKa = 9.81PEE282 pKa = 3.79RR283 pKa = 11.84TGRR286 pKa = 11.84FVLDD290 pKa = 3.6GALDD294 pKa = 3.78PRR296 pKa = 11.84LTADD300 pKa = 5.23DD301 pKa = 3.58ITRR304 pKa = 11.84GQAEE308 pKa = 4.46GFEE311 pKa = 4.5EE312 pKa = 4.12ATSAFVEE319 pKa = 4.66SCLSQGSDD327 pKa = 3.31GCPLTGSVEE336 pKa = 4.17DD337 pKa = 4.0GKK339 pKa = 10.85EE340 pKa = 3.62QLRR343 pKa = 11.84AFFDD347 pKa = 3.86SVDD350 pKa = 3.7AQPLQTDD357 pKa = 3.8DD358 pKa = 4.04PNRR361 pKa = 11.84PLTGALARR369 pKa = 11.84SGLLVVLYY377 pKa = 10.7NDD379 pKa = 3.96EE380 pKa = 4.09NWPLGVQALNAAMTGDD396 pKa = 3.57GSTLLFLADD405 pKa = 4.21LSADD409 pKa = 3.96RR410 pKa = 11.84EE411 pKa = 4.24DD412 pKa = 4.4DD413 pKa = 3.33GTYY416 pKa = 10.54SGNGTFAITAVNCLDD431 pKa = 3.64HH432 pKa = 8.2VGVADD437 pKa = 3.92QQWQIDD443 pKa = 3.77EE444 pKa = 4.3AAAVAEE450 pKa = 4.8EE451 pKa = 4.45FPTWGPGFGFSQTMCDD467 pKa = 3.5LWPAKK472 pKa = 9.42PVRR475 pKa = 11.84EE476 pKa = 4.08PAPISAAGSDD486 pKa = 4.28LIVVIGTRR494 pKa = 11.84GDD496 pKa = 3.38PATPYY501 pKa = 10.55DD502 pKa = 3.56WAVGLDD508 pKa = 3.6EE509 pKa = 4.28QLEE512 pKa = 4.22NSTLITYY519 pKa = 9.63EE520 pKa = 4.34GEE522 pKa = 3.66GHH524 pKa = 5.38TAYY527 pKa = 10.17GRR529 pKa = 11.84SGGCVEE535 pKa = 4.59AAVDD539 pKa = 4.26AYY541 pKa = 10.68LLSGTAPEE549 pKa = 4.62EE550 pKa = 4.25GLTCSS555 pKa = 3.97
Molecular weight: 57.68 kDa
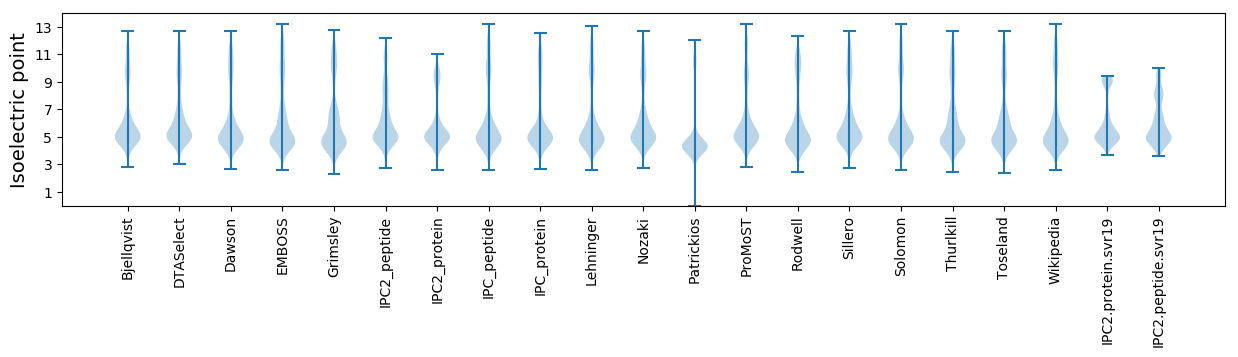
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Z9JQY5|Z9JQY5_9MICO Ribosome hibernation promoting factor OS=Brachybacterium phenoliresistens OX=396014 GN=hpf PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.1KK16 pKa = 8.61HH17 pKa = 4.4GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIINARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.68GRR40 pKa = 11.84AEE42 pKa = 4.11LSAA45 pKa = 4.93
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.1KK16 pKa = 8.61HH17 pKa = 4.4GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIINARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.68GRR40 pKa = 11.84AEE42 pKa = 4.11LSAA45 pKa = 4.93
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230214 |
37 |
2216 |
341.3 |
36.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.153 ± 0.06 | 0.565 ± 0.009 |
6.054 ± 0.037 | 6.159 ± 0.04 |
2.686 ± 0.023 | 9.336 ± 0.036 |
2.182 ± 0.021 | 4.092 ± 0.026 |
1.346 ± 0.025 | 10.6 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.914 ± 0.017 | 1.416 ± 0.019 |
6.109 ± 0.032 | 2.836 ± 0.021 |
7.982 ± 0.047 | 5.361 ± 0.03 |
5.654 ± 0.029 | 8.326 ± 0.037 |
1.455 ± 0.016 | 1.774 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
