
Black sea bass polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Centropristis striata polyomavirus 1
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
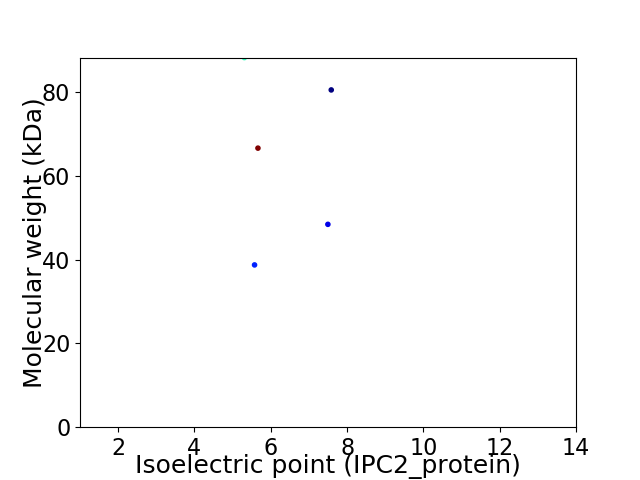
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1C723|A0A0A1C723_9POLY SLT OS=Black sea bass polyomavirus 1 OX=1572341 PE=4 SV=1
MM1 pKa = 7.59SEE3 pKa = 3.81QSNSFEE9 pKa = 3.97PRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84ALGEE18 pKa = 4.04HH19 pKa = 5.93NNPRR23 pKa = 11.84KK24 pKa = 9.72SVRR27 pKa = 11.84EE28 pKa = 3.87CWEE31 pKa = 3.61SWKK34 pKa = 10.94AEE36 pKa = 4.07QQKK39 pKa = 10.33KK40 pKa = 9.87PSLSEE45 pKa = 3.58EE46 pKa = 3.74DD47 pKa = 3.17RR48 pKa = 11.84GYY50 pKa = 10.83GDD52 pKa = 4.56SMVTLYY58 pKa = 10.35EE59 pKa = 3.86QYY61 pKa = 9.63TQVEE65 pKa = 4.51SALEE69 pKa = 4.1SFSASQTPEE78 pKa = 3.77KK79 pKa = 10.55VSPVQFDD86 pKa = 3.48EE87 pKa = 4.13PLRR90 pKa = 11.84EE91 pKa = 4.09FLCSYY96 pKa = 11.01AGFGHH101 pKa = 6.77SYY103 pKa = 10.98ILMCGEE109 pKa = 4.73EE110 pKa = 4.06KK111 pKa = 10.03WKK113 pKa = 10.45LYY115 pKa = 10.13QRR117 pKa = 11.84ILFTYY122 pKa = 7.9RR123 pKa = 11.84TEE125 pKa = 5.21DD126 pKa = 3.35RR127 pKa = 11.84FCLSRR132 pKa = 11.84PTPKK136 pKa = 9.79GTVYY140 pKa = 10.8YY141 pKa = 10.09ISIKK145 pKa = 8.07TRR147 pKa = 11.84YY148 pKa = 9.3RR149 pKa = 11.84IGGDD153 pKa = 3.63VVWNKK158 pKa = 9.67CRR160 pKa = 11.84AIGADD165 pKa = 3.09SWFEE169 pKa = 3.71LSAVKK174 pKa = 10.23VKK176 pKa = 10.45KK177 pKa = 9.88YY178 pKa = 9.99QQLKK182 pKa = 9.68ACAFGLGGVEE192 pKa = 3.68ITEE195 pKa = 4.31GEE197 pKa = 4.41LEE199 pKa = 4.23FEE201 pKa = 4.41RR202 pKa = 11.84PPFKK206 pKa = 10.74YY207 pKa = 10.42ALLEE211 pKa = 4.29KK212 pKa = 10.5YY213 pKa = 10.15AVEE216 pKa = 4.44TGCCDD221 pKa = 3.63PLLLLGDD228 pKa = 4.06YY229 pKa = 9.36LTFAEE234 pKa = 5.4GLQSCDD240 pKa = 2.47EE241 pKa = 4.77CYY243 pKa = 10.27QEE245 pKa = 4.28QLRR248 pKa = 11.84VADD251 pKa = 3.59PAGRR255 pKa = 11.84VHH257 pKa = 5.54STRR260 pKa = 11.84HH261 pKa = 4.96EE262 pKa = 3.95AHH264 pKa = 6.4HH265 pKa = 5.97QNAKK269 pKa = 9.78GFKK272 pKa = 9.78DD273 pKa = 3.58VSNKK277 pKa = 9.57KK278 pKa = 7.05QQCVSACDD286 pKa = 3.8VVCGCLRR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 4.3TKK298 pKa = 9.43MSRR301 pKa = 11.84DD302 pKa = 3.38DD303 pKa = 3.66RR304 pKa = 11.84FGARR308 pKa = 11.84VLQGLEE314 pKa = 3.73NLVRR318 pKa = 11.84GVTTEE323 pKa = 3.86MLSAAFLLKK332 pKa = 10.88AMLPFPITEE341 pKa = 4.29FFDD344 pKa = 4.09KK345 pKa = 10.81VVKK348 pKa = 10.06TMVEE352 pKa = 4.1AVPKK356 pKa = 8.69KK357 pKa = 9.97QGWVFRR363 pKa = 11.84GPIDD367 pKa = 3.59SGKK370 pKa = 8.59STVAAAVCGLLGGVALNVNTNRR392 pKa = 11.84DD393 pKa = 4.03RR394 pKa = 11.84LWVEE398 pKa = 3.92LGRR401 pKa = 11.84ALDD404 pKa = 3.46RR405 pKa = 11.84YY406 pKa = 8.75MVVFEE411 pKa = 4.98DD412 pKa = 3.63VKK414 pKa = 11.06GMPEE418 pKa = 3.87RR419 pKa = 11.84GHH421 pKa = 6.81PEE423 pKa = 3.81LPFGEE428 pKa = 4.77GFPNLDD434 pKa = 3.72SLRR437 pKa = 11.84EE438 pKa = 4.02CLDD441 pKa = 3.21GLFKK445 pKa = 11.11VGLEE449 pKa = 3.75RR450 pKa = 11.84KK451 pKa = 8.51HH452 pKa = 4.93QNKK455 pKa = 9.35VEE457 pKa = 4.21THH459 pKa = 5.76FPPWIVTCNQYY470 pKa = 10.53IIPEE474 pKa = 4.45TILARR479 pKa = 11.84CKK481 pKa = 10.83VLDD484 pKa = 4.07FAPGRR489 pKa = 11.84LNFKK493 pKa = 10.55QFVEE497 pKa = 4.24QKK499 pKa = 10.6GVDD502 pKa = 4.0LRR504 pKa = 11.84FMASGEE510 pKa = 4.24CLLLIYY516 pKa = 10.75CMFGDD521 pKa = 3.93LTLFQGEE528 pKa = 4.22QAVALSKK535 pKa = 10.65QVSDD539 pKa = 4.23EE540 pKa = 4.12AALLPWGLASEE551 pKa = 4.73GADD554 pKa = 3.51TQDD557 pKa = 4.79EE558 pKa = 4.32EE559 pKa = 5.02SQDD562 pKa = 3.7STLSTIPVEE571 pKa = 4.44CAPPSPGLRR580 pKa = 11.84LEE582 pKa = 4.31VRR584 pKa = 11.84GPPHH588 pKa = 5.54IRR590 pKa = 11.84VRR592 pKa = 11.84VRR594 pKa = 11.84VSEE597 pKa = 4.16PPAYY601 pKa = 9.62RR602 pKa = 11.84DD603 pKa = 3.47PPPYY607 pKa = 9.93PGPTGLQDD615 pKa = 3.64LCSTAFTDD623 pKa = 4.81DD624 pKa = 4.14PEE626 pKa = 5.26HH627 pKa = 6.65LFSPGPPPFSPPDD640 pKa = 3.77PGMHH644 pKa = 6.14QGDD647 pKa = 4.62SEE649 pKa = 4.66PEE651 pKa = 3.88SSPCEE656 pKa = 3.68MVSIAEE662 pKa = 4.01YY663 pKa = 10.4KK664 pKa = 9.74RR665 pKa = 11.84RR666 pKa = 11.84HH667 pKa = 5.95NIGSDD672 pKa = 3.17GLPNKK677 pKa = 9.81RR678 pKa = 11.84RR679 pKa = 11.84RR680 pKa = 11.84CLFVDD685 pKa = 5.81DD686 pKa = 5.04EE687 pKa = 4.62AQCTDD692 pKa = 3.82DD693 pKa = 3.61EE694 pKa = 4.89SEE696 pKa = 4.18EE697 pKa = 4.13EE698 pKa = 4.21EE699 pKa = 4.4EE700 pKa = 4.54EE701 pKa = 4.61EE702 pKa = 4.63EE703 pKa = 5.5DD704 pKa = 3.93EE705 pKa = 5.66QPSCSQRR712 pKa = 11.84PTTPVTPARR721 pKa = 11.84THH723 pKa = 5.29QRR725 pKa = 11.84PITDD729 pKa = 4.55FLRR732 pKa = 11.84QPLHH736 pKa = 6.75PEE738 pKa = 4.24GSKK741 pKa = 10.84CPYY744 pKa = 9.9KK745 pKa = 10.75DD746 pKa = 3.09AGCFFDD752 pKa = 4.69HH753 pKa = 7.48PVRR756 pKa = 11.84PWCDD760 pKa = 2.47RR761 pKa = 11.84CLRR764 pKa = 11.84DD765 pKa = 2.94WVTPEE770 pKa = 3.6KK771 pKa = 10.65VPRR774 pKa = 11.84PGTPTPPSVV783 pKa = 3.03
MM1 pKa = 7.59SEE3 pKa = 3.81QSNSFEE9 pKa = 3.97PRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84ALGEE18 pKa = 4.04HH19 pKa = 5.93NNPRR23 pKa = 11.84KK24 pKa = 9.72SVRR27 pKa = 11.84EE28 pKa = 3.87CWEE31 pKa = 3.61SWKK34 pKa = 10.94AEE36 pKa = 4.07QQKK39 pKa = 10.33KK40 pKa = 9.87PSLSEE45 pKa = 3.58EE46 pKa = 3.74DD47 pKa = 3.17RR48 pKa = 11.84GYY50 pKa = 10.83GDD52 pKa = 4.56SMVTLYY58 pKa = 10.35EE59 pKa = 3.86QYY61 pKa = 9.63TQVEE65 pKa = 4.51SALEE69 pKa = 4.1SFSASQTPEE78 pKa = 3.77KK79 pKa = 10.55VSPVQFDD86 pKa = 3.48EE87 pKa = 4.13PLRR90 pKa = 11.84EE91 pKa = 4.09FLCSYY96 pKa = 11.01AGFGHH101 pKa = 6.77SYY103 pKa = 10.98ILMCGEE109 pKa = 4.73EE110 pKa = 4.06KK111 pKa = 10.03WKK113 pKa = 10.45LYY115 pKa = 10.13QRR117 pKa = 11.84ILFTYY122 pKa = 7.9RR123 pKa = 11.84TEE125 pKa = 5.21DD126 pKa = 3.35RR127 pKa = 11.84FCLSRR132 pKa = 11.84PTPKK136 pKa = 9.79GTVYY140 pKa = 10.8YY141 pKa = 10.09ISIKK145 pKa = 8.07TRR147 pKa = 11.84YY148 pKa = 9.3RR149 pKa = 11.84IGGDD153 pKa = 3.63VVWNKK158 pKa = 9.67CRR160 pKa = 11.84AIGADD165 pKa = 3.09SWFEE169 pKa = 3.71LSAVKK174 pKa = 10.23VKK176 pKa = 10.45KK177 pKa = 9.88YY178 pKa = 9.99QQLKK182 pKa = 9.68ACAFGLGGVEE192 pKa = 3.68ITEE195 pKa = 4.31GEE197 pKa = 4.41LEE199 pKa = 4.23FEE201 pKa = 4.41RR202 pKa = 11.84PPFKK206 pKa = 10.74YY207 pKa = 10.42ALLEE211 pKa = 4.29KK212 pKa = 10.5YY213 pKa = 10.15AVEE216 pKa = 4.44TGCCDD221 pKa = 3.63PLLLLGDD228 pKa = 4.06YY229 pKa = 9.36LTFAEE234 pKa = 5.4GLQSCDD240 pKa = 2.47EE241 pKa = 4.77CYY243 pKa = 10.27QEE245 pKa = 4.28QLRR248 pKa = 11.84VADD251 pKa = 3.59PAGRR255 pKa = 11.84VHH257 pKa = 5.54STRR260 pKa = 11.84HH261 pKa = 4.96EE262 pKa = 3.95AHH264 pKa = 6.4HH265 pKa = 5.97QNAKK269 pKa = 9.78GFKK272 pKa = 9.78DD273 pKa = 3.58VSNKK277 pKa = 9.57KK278 pKa = 7.05QQCVSACDD286 pKa = 3.8VVCGCLRR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 4.3TKK298 pKa = 9.43MSRR301 pKa = 11.84DD302 pKa = 3.38DD303 pKa = 3.66RR304 pKa = 11.84FGARR308 pKa = 11.84VLQGLEE314 pKa = 3.73NLVRR318 pKa = 11.84GVTTEE323 pKa = 3.86MLSAAFLLKK332 pKa = 10.88AMLPFPITEE341 pKa = 4.29FFDD344 pKa = 4.09KK345 pKa = 10.81VVKK348 pKa = 10.06TMVEE352 pKa = 4.1AVPKK356 pKa = 8.69KK357 pKa = 9.97QGWVFRR363 pKa = 11.84GPIDD367 pKa = 3.59SGKK370 pKa = 8.59STVAAAVCGLLGGVALNVNTNRR392 pKa = 11.84DD393 pKa = 4.03RR394 pKa = 11.84LWVEE398 pKa = 3.92LGRR401 pKa = 11.84ALDD404 pKa = 3.46RR405 pKa = 11.84YY406 pKa = 8.75MVVFEE411 pKa = 4.98DD412 pKa = 3.63VKK414 pKa = 11.06GMPEE418 pKa = 3.87RR419 pKa = 11.84GHH421 pKa = 6.81PEE423 pKa = 3.81LPFGEE428 pKa = 4.77GFPNLDD434 pKa = 3.72SLRR437 pKa = 11.84EE438 pKa = 4.02CLDD441 pKa = 3.21GLFKK445 pKa = 11.11VGLEE449 pKa = 3.75RR450 pKa = 11.84KK451 pKa = 8.51HH452 pKa = 4.93QNKK455 pKa = 9.35VEE457 pKa = 4.21THH459 pKa = 5.76FPPWIVTCNQYY470 pKa = 10.53IIPEE474 pKa = 4.45TILARR479 pKa = 11.84CKK481 pKa = 10.83VLDD484 pKa = 4.07FAPGRR489 pKa = 11.84LNFKK493 pKa = 10.55QFVEE497 pKa = 4.24QKK499 pKa = 10.6GVDD502 pKa = 4.0LRR504 pKa = 11.84FMASGEE510 pKa = 4.24CLLLIYY516 pKa = 10.75CMFGDD521 pKa = 3.93LTLFQGEE528 pKa = 4.22QAVALSKK535 pKa = 10.65QVSDD539 pKa = 4.23EE540 pKa = 4.12AALLPWGLASEE551 pKa = 4.73GADD554 pKa = 3.51TQDD557 pKa = 4.79EE558 pKa = 4.32EE559 pKa = 5.02SQDD562 pKa = 3.7STLSTIPVEE571 pKa = 4.44CAPPSPGLRR580 pKa = 11.84LEE582 pKa = 4.31VRR584 pKa = 11.84GPPHH588 pKa = 5.54IRR590 pKa = 11.84VRR592 pKa = 11.84VRR594 pKa = 11.84VSEE597 pKa = 4.16PPAYY601 pKa = 9.62RR602 pKa = 11.84DD603 pKa = 3.47PPPYY607 pKa = 9.93PGPTGLQDD615 pKa = 3.64LCSTAFTDD623 pKa = 4.81DD624 pKa = 4.14PEE626 pKa = 5.26HH627 pKa = 6.65LFSPGPPPFSPPDD640 pKa = 3.77PGMHH644 pKa = 6.14QGDD647 pKa = 4.62SEE649 pKa = 4.66PEE651 pKa = 3.88SSPCEE656 pKa = 3.68MVSIAEE662 pKa = 4.01YY663 pKa = 10.4KK664 pKa = 9.74RR665 pKa = 11.84RR666 pKa = 11.84HH667 pKa = 5.95NIGSDD672 pKa = 3.17GLPNKK677 pKa = 9.81RR678 pKa = 11.84RR679 pKa = 11.84RR680 pKa = 11.84CLFVDD685 pKa = 5.81DD686 pKa = 5.04EE687 pKa = 4.62AQCTDD692 pKa = 3.82DD693 pKa = 3.61EE694 pKa = 4.89SEE696 pKa = 4.18EE697 pKa = 4.13EE698 pKa = 4.21EE699 pKa = 4.4EE700 pKa = 4.54EE701 pKa = 4.61EE702 pKa = 4.63EE703 pKa = 5.5DD704 pKa = 3.93EE705 pKa = 5.66QPSCSQRR712 pKa = 11.84PTTPVTPARR721 pKa = 11.84THH723 pKa = 5.29QRR725 pKa = 11.84PITDD729 pKa = 4.55FLRR732 pKa = 11.84QPLHH736 pKa = 6.75PEE738 pKa = 4.24GSKK741 pKa = 10.84CPYY744 pKa = 9.9KK745 pKa = 10.75DD746 pKa = 3.09AGCFFDD752 pKa = 4.69HH753 pKa = 7.48PVRR756 pKa = 11.84PWCDD760 pKa = 2.47RR761 pKa = 11.84CLRR764 pKa = 11.84DD765 pKa = 2.94WVTPEE770 pKa = 3.6KK771 pKa = 10.65VPRR774 pKa = 11.84PGTPTPPSVV783 pKa = 3.03
Molecular weight: 88.31 kDa
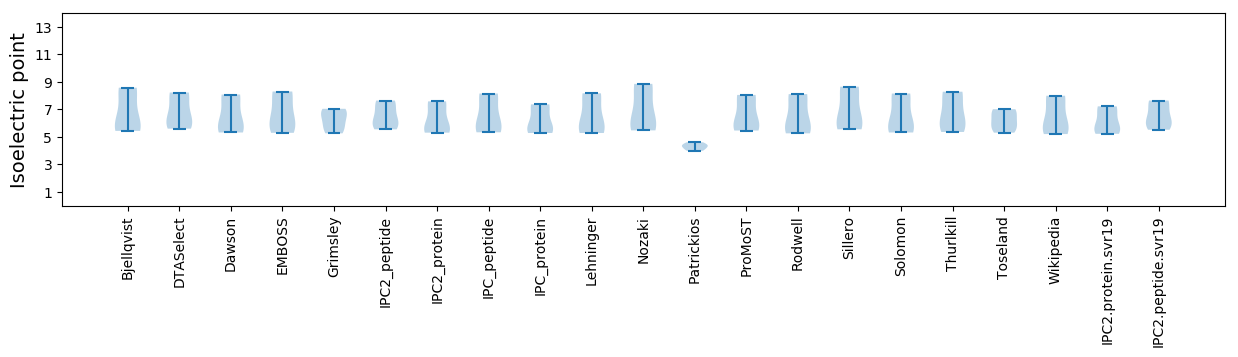
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1C950|A0A0A1C950_9POLY VP2 OS=Black sea bass polyomavirus 1 OX=1572341 GN=VP2 PE=4 SV=1
MM1 pKa = 7.59SEE3 pKa = 3.81QSNSFEE9 pKa = 3.97PRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84ALGEE18 pKa = 4.04HH19 pKa = 5.93NNPRR23 pKa = 11.84KK24 pKa = 9.72SVRR27 pKa = 11.84EE28 pKa = 3.87CWEE31 pKa = 3.61SWKK34 pKa = 10.94AEE36 pKa = 4.07QQKK39 pKa = 10.33KK40 pKa = 9.87PSLSEE45 pKa = 3.58EE46 pKa = 3.74DD47 pKa = 3.17RR48 pKa = 11.84GYY50 pKa = 10.83GDD52 pKa = 4.56SMVTLYY58 pKa = 10.35EE59 pKa = 3.86QYY61 pKa = 9.63TQVEE65 pKa = 4.51SALEE69 pKa = 4.1SFSASQTPEE78 pKa = 3.77KK79 pKa = 10.55VSPVQFDD86 pKa = 3.48EE87 pKa = 4.13PLRR90 pKa = 11.84EE91 pKa = 4.09FLCSYY96 pKa = 11.01AGFGHH101 pKa = 6.77SYY103 pKa = 10.98ILMCGEE109 pKa = 4.73EE110 pKa = 4.06KK111 pKa = 10.03WKK113 pKa = 10.45LYY115 pKa = 10.13QRR117 pKa = 11.84ILFTYY122 pKa = 7.9RR123 pKa = 11.84TEE125 pKa = 5.21DD126 pKa = 3.35RR127 pKa = 11.84FCLSRR132 pKa = 11.84PTPKK136 pKa = 9.79GTVYY140 pKa = 10.8YY141 pKa = 10.09ISIKK145 pKa = 8.07TRR147 pKa = 11.84YY148 pKa = 9.3RR149 pKa = 11.84IGGDD153 pKa = 3.63VVWNKK158 pKa = 9.67CRR160 pKa = 11.84AIGADD165 pKa = 3.09SWFEE169 pKa = 3.71LSAVKK174 pKa = 10.23VKK176 pKa = 10.45KK177 pKa = 9.88YY178 pKa = 9.99QQLKK182 pKa = 9.68ACAFGLGGVEE192 pKa = 3.68ITEE195 pKa = 4.31GEE197 pKa = 4.41LEE199 pKa = 4.23FEE201 pKa = 4.41RR202 pKa = 11.84PPFKK206 pKa = 10.74YY207 pKa = 10.42ALLEE211 pKa = 4.29KK212 pKa = 10.5YY213 pKa = 10.15AVEE216 pKa = 4.44TGCCDD221 pKa = 3.63PLLLLGDD228 pKa = 4.06YY229 pKa = 9.36LTFAEE234 pKa = 5.4GLQSCDD240 pKa = 2.47EE241 pKa = 4.77CYY243 pKa = 10.27QEE245 pKa = 4.28QLRR248 pKa = 11.84VADD251 pKa = 3.59PAGRR255 pKa = 11.84VHH257 pKa = 5.54STRR260 pKa = 11.84HH261 pKa = 4.96EE262 pKa = 3.95AHH264 pKa = 6.4HH265 pKa = 5.97QNAKK269 pKa = 9.78GFKK272 pKa = 9.78DD273 pKa = 3.58VSNKK277 pKa = 9.57KK278 pKa = 7.05QQCVSACDD286 pKa = 3.8VVCGCLRR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 4.3TKK298 pKa = 9.43MSRR301 pKa = 11.84DD302 pKa = 3.38DD303 pKa = 3.66RR304 pKa = 11.84FGARR308 pKa = 11.84VLQGLEE314 pKa = 3.73NLVRR318 pKa = 11.84GVTTEE323 pKa = 3.86MLSAAFLLKK332 pKa = 10.88AMLPFPITEE341 pKa = 4.29FFDD344 pKa = 4.09KK345 pKa = 10.81VVKK348 pKa = 10.06TMVEE352 pKa = 4.1AVPKK356 pKa = 8.69KK357 pKa = 9.97QGWVFRR363 pKa = 11.84GPIDD367 pKa = 3.59SGKK370 pKa = 8.59STVAAAVCGLLGGVALNVNTNRR392 pKa = 11.84DD393 pKa = 4.03RR394 pKa = 11.84LWVEE398 pKa = 3.92LGRR401 pKa = 11.84ALDD404 pKa = 3.46RR405 pKa = 11.84YY406 pKa = 8.75MVVFEE411 pKa = 4.98DD412 pKa = 3.63VKK414 pKa = 11.06GMPEE418 pKa = 3.87RR419 pKa = 11.84GHH421 pKa = 6.81PEE423 pKa = 3.81LPFGEE428 pKa = 4.77GFPNLDD434 pKa = 3.72SLRR437 pKa = 11.84EE438 pKa = 4.02CLDD441 pKa = 3.21GLFKK445 pKa = 11.11VGLEE449 pKa = 3.75RR450 pKa = 11.84KK451 pKa = 8.51HH452 pKa = 4.93QNKK455 pKa = 9.35VEE457 pKa = 4.21THH459 pKa = 5.76FPPWIVTCNQYY470 pKa = 10.53IIPEE474 pKa = 4.45TILARR479 pKa = 11.84CKK481 pKa = 10.83VLDD484 pKa = 4.07FAPGRR489 pKa = 11.84LNFKK493 pKa = 10.55QFVEE497 pKa = 4.24QKK499 pKa = 10.6GVDD502 pKa = 4.0LRR504 pKa = 11.84FMASGEE510 pKa = 4.24CLLLIYY516 pKa = 10.75CMFGDD521 pKa = 3.93LTLFQGEE528 pKa = 4.22QAVALSKK535 pKa = 10.65QVSDD539 pKa = 4.23EE540 pKa = 4.12AALLPWGLASEE551 pKa = 4.73GADD554 pKa = 3.51TQDD557 pKa = 4.79EE558 pKa = 4.32EE559 pKa = 5.02SQDD562 pKa = 3.7STLSTIPVEE571 pKa = 4.44CAPPSPGLRR580 pKa = 11.84LEE582 pKa = 4.31LSLMILNIFFLLVPPLFLLLILACIRR608 pKa = 11.84ATLNLSRR615 pKa = 11.84HH616 pKa = 4.99PVRR619 pKa = 11.84WSPSQSTSAAIILGQMVYY637 pKa = 9.68PINVGVVSSWMTRR650 pKa = 11.84HH651 pKa = 5.95SAPMMRR657 pKa = 11.84ARR659 pKa = 11.84RR660 pKa = 11.84KK661 pKa = 8.29RR662 pKa = 11.84RR663 pKa = 11.84RR664 pKa = 11.84RR665 pKa = 11.84RR666 pKa = 11.84TSNPVVPSVPPPQSPLPGPTNVLSRR691 pKa = 11.84IFSVSLSTLKK701 pKa = 10.85GPSVLIRR708 pKa = 11.84TRR710 pKa = 11.84AASLTTPP717 pKa = 3.89
MM1 pKa = 7.59SEE3 pKa = 3.81QSNSFEE9 pKa = 3.97PRR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84ALGEE18 pKa = 4.04HH19 pKa = 5.93NNPRR23 pKa = 11.84KK24 pKa = 9.72SVRR27 pKa = 11.84EE28 pKa = 3.87CWEE31 pKa = 3.61SWKK34 pKa = 10.94AEE36 pKa = 4.07QQKK39 pKa = 10.33KK40 pKa = 9.87PSLSEE45 pKa = 3.58EE46 pKa = 3.74DD47 pKa = 3.17RR48 pKa = 11.84GYY50 pKa = 10.83GDD52 pKa = 4.56SMVTLYY58 pKa = 10.35EE59 pKa = 3.86QYY61 pKa = 9.63TQVEE65 pKa = 4.51SALEE69 pKa = 4.1SFSASQTPEE78 pKa = 3.77KK79 pKa = 10.55VSPVQFDD86 pKa = 3.48EE87 pKa = 4.13PLRR90 pKa = 11.84EE91 pKa = 4.09FLCSYY96 pKa = 11.01AGFGHH101 pKa = 6.77SYY103 pKa = 10.98ILMCGEE109 pKa = 4.73EE110 pKa = 4.06KK111 pKa = 10.03WKK113 pKa = 10.45LYY115 pKa = 10.13QRR117 pKa = 11.84ILFTYY122 pKa = 7.9RR123 pKa = 11.84TEE125 pKa = 5.21DD126 pKa = 3.35RR127 pKa = 11.84FCLSRR132 pKa = 11.84PTPKK136 pKa = 9.79GTVYY140 pKa = 10.8YY141 pKa = 10.09ISIKK145 pKa = 8.07TRR147 pKa = 11.84YY148 pKa = 9.3RR149 pKa = 11.84IGGDD153 pKa = 3.63VVWNKK158 pKa = 9.67CRR160 pKa = 11.84AIGADD165 pKa = 3.09SWFEE169 pKa = 3.71LSAVKK174 pKa = 10.23VKK176 pKa = 10.45KK177 pKa = 9.88YY178 pKa = 9.99QQLKK182 pKa = 9.68ACAFGLGGVEE192 pKa = 3.68ITEE195 pKa = 4.31GEE197 pKa = 4.41LEE199 pKa = 4.23FEE201 pKa = 4.41RR202 pKa = 11.84PPFKK206 pKa = 10.74YY207 pKa = 10.42ALLEE211 pKa = 4.29KK212 pKa = 10.5YY213 pKa = 10.15AVEE216 pKa = 4.44TGCCDD221 pKa = 3.63PLLLLGDD228 pKa = 4.06YY229 pKa = 9.36LTFAEE234 pKa = 5.4GLQSCDD240 pKa = 2.47EE241 pKa = 4.77CYY243 pKa = 10.27QEE245 pKa = 4.28QLRR248 pKa = 11.84VADD251 pKa = 3.59PAGRR255 pKa = 11.84VHH257 pKa = 5.54STRR260 pKa = 11.84HH261 pKa = 4.96EE262 pKa = 3.95AHH264 pKa = 6.4HH265 pKa = 5.97QNAKK269 pKa = 9.78GFKK272 pKa = 9.78DD273 pKa = 3.58VSNKK277 pKa = 9.57KK278 pKa = 7.05QQCVSACDD286 pKa = 3.8VVCGCLRR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 4.3TKK298 pKa = 9.43MSRR301 pKa = 11.84DD302 pKa = 3.38DD303 pKa = 3.66RR304 pKa = 11.84FGARR308 pKa = 11.84VLQGLEE314 pKa = 3.73NLVRR318 pKa = 11.84GVTTEE323 pKa = 3.86MLSAAFLLKK332 pKa = 10.88AMLPFPITEE341 pKa = 4.29FFDD344 pKa = 4.09KK345 pKa = 10.81VVKK348 pKa = 10.06TMVEE352 pKa = 4.1AVPKK356 pKa = 8.69KK357 pKa = 9.97QGWVFRR363 pKa = 11.84GPIDD367 pKa = 3.59SGKK370 pKa = 8.59STVAAAVCGLLGGVALNVNTNRR392 pKa = 11.84DD393 pKa = 4.03RR394 pKa = 11.84LWVEE398 pKa = 3.92LGRR401 pKa = 11.84ALDD404 pKa = 3.46RR405 pKa = 11.84YY406 pKa = 8.75MVVFEE411 pKa = 4.98DD412 pKa = 3.63VKK414 pKa = 11.06GMPEE418 pKa = 3.87RR419 pKa = 11.84GHH421 pKa = 6.81PEE423 pKa = 3.81LPFGEE428 pKa = 4.77GFPNLDD434 pKa = 3.72SLRR437 pKa = 11.84EE438 pKa = 4.02CLDD441 pKa = 3.21GLFKK445 pKa = 11.11VGLEE449 pKa = 3.75RR450 pKa = 11.84KK451 pKa = 8.51HH452 pKa = 4.93QNKK455 pKa = 9.35VEE457 pKa = 4.21THH459 pKa = 5.76FPPWIVTCNQYY470 pKa = 10.53IIPEE474 pKa = 4.45TILARR479 pKa = 11.84CKK481 pKa = 10.83VLDD484 pKa = 4.07FAPGRR489 pKa = 11.84LNFKK493 pKa = 10.55QFVEE497 pKa = 4.24QKK499 pKa = 10.6GVDD502 pKa = 4.0LRR504 pKa = 11.84FMASGEE510 pKa = 4.24CLLLIYY516 pKa = 10.75CMFGDD521 pKa = 3.93LTLFQGEE528 pKa = 4.22QAVALSKK535 pKa = 10.65QVSDD539 pKa = 4.23EE540 pKa = 4.12AALLPWGLASEE551 pKa = 4.73GADD554 pKa = 3.51TQDD557 pKa = 4.79EE558 pKa = 4.32EE559 pKa = 5.02SQDD562 pKa = 3.7STLSTIPVEE571 pKa = 4.44CAPPSPGLRR580 pKa = 11.84LEE582 pKa = 4.31LSLMILNIFFLLVPPLFLLLILACIRR608 pKa = 11.84ATLNLSRR615 pKa = 11.84HH616 pKa = 4.99PVRR619 pKa = 11.84WSPSQSTSAAIILGQMVYY637 pKa = 9.68PINVGVVSSWMTRR650 pKa = 11.84HH651 pKa = 5.95SAPMMRR657 pKa = 11.84ARR659 pKa = 11.84RR660 pKa = 11.84KK661 pKa = 8.29RR662 pKa = 11.84RR663 pKa = 11.84RR664 pKa = 11.84RR665 pKa = 11.84RR666 pKa = 11.84TSNPVVPSVPPPQSPLPGPTNVLSRR691 pKa = 11.84IFSVSLSTLKK701 pKa = 10.85GPSVLIRR708 pKa = 11.84TRR710 pKa = 11.84AASLTTPP717 pKa = 3.89
Molecular weight: 80.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2889 |
353 |
783 |
577.8 |
64.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.784 ± 1.347 | 2.25 ± 0.611 |
5.365 ± 0.671 | 7.58 ± 0.624 |
4.05 ± 0.432 | 7.823 ± 0.548 |
1.904 ± 0.253 | 3.15 ± 0.248 |
5.85 ± 0.304 | 7.996 ± 0.874 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.355 | 2.873 ± 0.451 |
7.061 ± 0.764 | 4.361 ± 0.158 |
6.3 ± 0.771 | 7.2 ± 0.57 |
5.123 ± 0.292 | 7.13 ± 0.158 |
1.869 ± 0.186 | 2.838 ± 0.088 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
