
Air potato virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus; Air potato ampelovirus 1
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
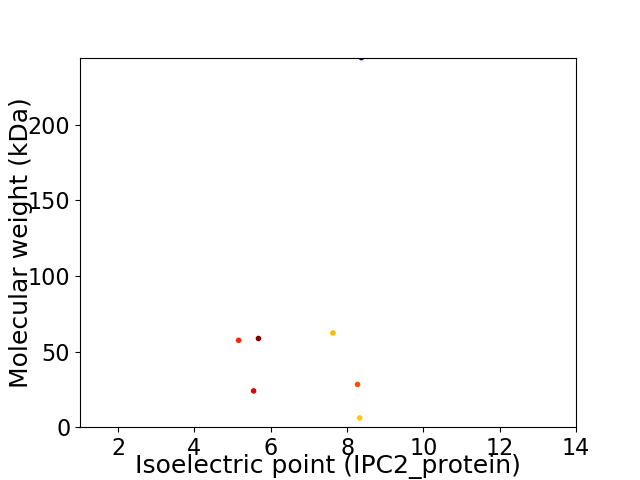
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G6V9C3|A0A3G6V9C3_9CLOS ORF5 OS=Air potato virus 1 OX=2491018 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.49SHH4 pKa = 6.95LEE6 pKa = 4.07VVNVMFDD13 pKa = 3.45HH14 pKa = 5.76YY15 pKa = 10.68QINGLPFSMKK25 pKa = 9.17WVEE28 pKa = 4.91DD29 pKa = 3.58LFQFSDD35 pKa = 3.93FSTVVDD41 pKa = 3.44NVTITEE47 pKa = 4.18CQKK50 pKa = 9.6TPKK53 pKa = 10.07AYY55 pKa = 9.58KK56 pKa = 9.89IKK58 pKa = 10.93SLVPLVNSLRR68 pKa = 11.84PSPRR72 pKa = 11.84PQSLISNLYY81 pKa = 9.09VFEE84 pKa = 4.13TRR86 pKa = 11.84NYY88 pKa = 8.56NADD91 pKa = 3.54RR92 pKa = 11.84GVQNYY97 pKa = 7.49YY98 pKa = 10.16RR99 pKa = 11.84PPVVKK104 pKa = 10.21EE105 pKa = 3.72LVDD108 pKa = 3.84TFFSTYY114 pKa = 9.23IDD116 pKa = 3.6AGRR119 pKa = 11.84MCEE122 pKa = 4.09MIGDD126 pKa = 3.64IRR128 pKa = 11.84VVNSVSLEE136 pKa = 3.73EE137 pKa = 4.35WLDD140 pKa = 3.31SRR142 pKa = 11.84PAMGKK147 pKa = 9.89SGLLNDD153 pKa = 4.09LTKK156 pKa = 10.95DD157 pKa = 3.28PYY159 pKa = 11.16YY160 pKa = 10.51QDD162 pKa = 3.44SLNHH166 pKa = 6.39FKK168 pKa = 11.46YY169 pKa = 9.68MIKK172 pKa = 10.29SDD174 pKa = 4.14LKK176 pKa = 11.24SKK178 pKa = 10.8LDD180 pKa = 3.35YY181 pKa = 9.61TARR184 pKa = 11.84EE185 pKa = 4.04NLASGQNIVYY195 pKa = 9.92HH196 pKa = 5.68EE197 pKa = 4.15RR198 pKa = 11.84RR199 pKa = 11.84ICALFSSVFQDD210 pKa = 3.36LVRR213 pKa = 11.84VLKK216 pKa = 10.49FVLADD221 pKa = 3.54KK222 pKa = 10.58FLLYY226 pKa = 10.61HH227 pKa = 6.47GVNLDD232 pKa = 3.55EE233 pKa = 5.18FAQAMTNKK241 pKa = 7.72MTFPLASCYY250 pKa = 10.25CGEE253 pKa = 4.12VDD255 pKa = 3.66ISKK258 pKa = 10.61YY259 pKa = 10.67DD260 pKa = 3.33KK261 pKa = 10.96SQNQMTKK268 pKa = 9.12EE269 pKa = 4.04VEE271 pKa = 3.81LEE273 pKa = 3.57IYY275 pKa = 10.02RR276 pKa = 11.84RR277 pKa = 11.84LGMHH281 pKa = 7.48PDD283 pKa = 4.37LVDD286 pKa = 3.95LWACSDD292 pKa = 3.75FSSRR296 pKa = 11.84VGSMKK301 pKa = 10.5EE302 pKa = 4.18GILLDD307 pKa = 3.24IGAQRR312 pKa = 11.84RR313 pKa = 11.84TGTATTWIGNTLINMTLLAAAGDD336 pKa = 3.86VKK338 pKa = 11.21NFDD341 pKa = 3.31IAAFSGDD348 pKa = 3.78DD349 pKa = 3.39SLLLSRR355 pKa = 11.84EE356 pKa = 4.04RR357 pKa = 11.84IEE359 pKa = 4.94LDD361 pKa = 3.24SFVYY365 pKa = 9.92EE366 pKa = 3.75YY367 pKa = 11.27HH368 pKa = 6.71FGFDD372 pKa = 3.26VKK374 pKa = 10.53FYY376 pKa = 10.98GQATPYY382 pKa = 9.71FCSKK386 pKa = 10.53FLIPVNGRR394 pKa = 11.84VVFVPDD400 pKa = 3.59ALKK403 pKa = 11.15LFVNLGAEE411 pKa = 4.09WDD413 pKa = 3.75CDD415 pKa = 3.77KK416 pKa = 11.94NEE418 pKa = 3.96MFEE421 pKa = 4.18KK422 pKa = 10.59FKK424 pKa = 11.44SFVDD428 pKa = 3.86LTKK431 pKa = 11.07DD432 pKa = 3.26LVDD435 pKa = 3.55NSVVEE440 pKa = 3.84ILAVYY445 pKa = 10.2IEE447 pKa = 4.53EE448 pKa = 4.9KK449 pKa = 10.57YY450 pKa = 11.09GSNPWVFPSLCCINSLRR467 pKa = 11.84SNFAQFMRR475 pKa = 11.84CWRR478 pKa = 11.84LCQKK482 pKa = 10.81LLLKK486 pKa = 10.59DD487 pKa = 3.89RR488 pKa = 11.84EE489 pKa = 4.34LGDD492 pKa = 3.46VAEE495 pKa = 5.88DD496 pKa = 3.43IDD498 pKa = 4.26EE499 pKa = 4.66GGG501 pKa = 3.31
MM1 pKa = 7.6KK2 pKa = 10.49SHH4 pKa = 6.95LEE6 pKa = 4.07VVNVMFDD13 pKa = 3.45HH14 pKa = 5.76YY15 pKa = 10.68QINGLPFSMKK25 pKa = 9.17WVEE28 pKa = 4.91DD29 pKa = 3.58LFQFSDD35 pKa = 3.93FSTVVDD41 pKa = 3.44NVTITEE47 pKa = 4.18CQKK50 pKa = 9.6TPKK53 pKa = 10.07AYY55 pKa = 9.58KK56 pKa = 9.89IKK58 pKa = 10.93SLVPLVNSLRR68 pKa = 11.84PSPRR72 pKa = 11.84PQSLISNLYY81 pKa = 9.09VFEE84 pKa = 4.13TRR86 pKa = 11.84NYY88 pKa = 8.56NADD91 pKa = 3.54RR92 pKa = 11.84GVQNYY97 pKa = 7.49YY98 pKa = 10.16RR99 pKa = 11.84PPVVKK104 pKa = 10.21EE105 pKa = 3.72LVDD108 pKa = 3.84TFFSTYY114 pKa = 9.23IDD116 pKa = 3.6AGRR119 pKa = 11.84MCEE122 pKa = 4.09MIGDD126 pKa = 3.64IRR128 pKa = 11.84VVNSVSLEE136 pKa = 3.73EE137 pKa = 4.35WLDD140 pKa = 3.31SRR142 pKa = 11.84PAMGKK147 pKa = 9.89SGLLNDD153 pKa = 4.09LTKK156 pKa = 10.95DD157 pKa = 3.28PYY159 pKa = 11.16YY160 pKa = 10.51QDD162 pKa = 3.44SLNHH166 pKa = 6.39FKK168 pKa = 11.46YY169 pKa = 9.68MIKK172 pKa = 10.29SDD174 pKa = 4.14LKK176 pKa = 11.24SKK178 pKa = 10.8LDD180 pKa = 3.35YY181 pKa = 9.61TARR184 pKa = 11.84EE185 pKa = 4.04NLASGQNIVYY195 pKa = 9.92HH196 pKa = 5.68EE197 pKa = 4.15RR198 pKa = 11.84RR199 pKa = 11.84ICALFSSVFQDD210 pKa = 3.36LVRR213 pKa = 11.84VLKK216 pKa = 10.49FVLADD221 pKa = 3.54KK222 pKa = 10.58FLLYY226 pKa = 10.61HH227 pKa = 6.47GVNLDD232 pKa = 3.55EE233 pKa = 5.18FAQAMTNKK241 pKa = 7.72MTFPLASCYY250 pKa = 10.25CGEE253 pKa = 4.12VDD255 pKa = 3.66ISKK258 pKa = 10.61YY259 pKa = 10.67DD260 pKa = 3.33KK261 pKa = 10.96SQNQMTKK268 pKa = 9.12EE269 pKa = 4.04VEE271 pKa = 3.81LEE273 pKa = 3.57IYY275 pKa = 10.02RR276 pKa = 11.84RR277 pKa = 11.84LGMHH281 pKa = 7.48PDD283 pKa = 4.37LVDD286 pKa = 3.95LWACSDD292 pKa = 3.75FSSRR296 pKa = 11.84VGSMKK301 pKa = 10.5EE302 pKa = 4.18GILLDD307 pKa = 3.24IGAQRR312 pKa = 11.84RR313 pKa = 11.84TGTATTWIGNTLINMTLLAAAGDD336 pKa = 3.86VKK338 pKa = 11.21NFDD341 pKa = 3.31IAAFSGDD348 pKa = 3.78DD349 pKa = 3.39SLLLSRR355 pKa = 11.84EE356 pKa = 4.04RR357 pKa = 11.84IEE359 pKa = 4.94LDD361 pKa = 3.24SFVYY365 pKa = 9.92EE366 pKa = 3.75YY367 pKa = 11.27HH368 pKa = 6.71FGFDD372 pKa = 3.26VKK374 pKa = 10.53FYY376 pKa = 10.98GQATPYY382 pKa = 9.71FCSKK386 pKa = 10.53FLIPVNGRR394 pKa = 11.84VVFVPDD400 pKa = 3.59ALKK403 pKa = 11.15LFVNLGAEE411 pKa = 4.09WDD413 pKa = 3.75CDD415 pKa = 3.77KK416 pKa = 11.94NEE418 pKa = 3.96MFEE421 pKa = 4.18KK422 pKa = 10.59FKK424 pKa = 11.44SFVDD428 pKa = 3.86LTKK431 pKa = 11.07DD432 pKa = 3.26LVDD435 pKa = 3.55NSVVEE440 pKa = 3.84ILAVYY445 pKa = 10.2IEE447 pKa = 4.53EE448 pKa = 4.9KK449 pKa = 10.57YY450 pKa = 11.09GSNPWVFPSLCCINSLRR467 pKa = 11.84SNFAQFMRR475 pKa = 11.84CWRR478 pKa = 11.84LCQKK482 pKa = 10.81LLLKK486 pKa = 10.59DD487 pKa = 3.89RR488 pKa = 11.84EE489 pKa = 4.34LGDD492 pKa = 3.46VAEE495 pKa = 5.88DD496 pKa = 3.43IDD498 pKa = 4.26EE499 pKa = 4.66GGG501 pKa = 3.31
Molecular weight: 57.45 kDa
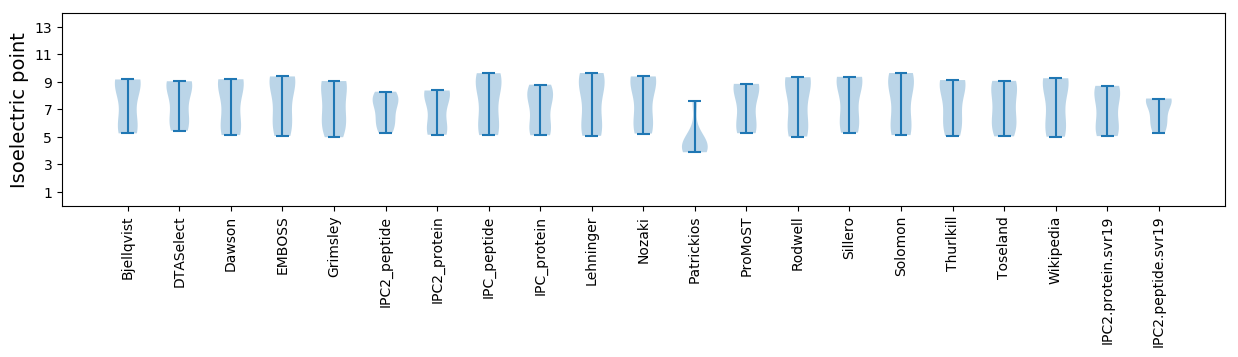
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G6V9B9|A0A3G6V9B9_9CLOS ORF3 OS=Air potato virus 1 OX=2491018 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.42VDD4 pKa = 4.25DD5 pKa = 4.39SVNTGVSFSIIVFTFFLLIVIAATVVFCLYY35 pKa = 10.78FLFTKK40 pKa = 10.46RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 3.71QSGVEE49 pKa = 3.76RR50 pKa = 11.84PTFF53 pKa = 3.54
MM1 pKa = 7.44KK2 pKa = 10.42VDD4 pKa = 4.25DD5 pKa = 4.39SVNTGVSFSIIVFTFFLLIVIAATVVFCLYY35 pKa = 10.78FLFTKK40 pKa = 10.46RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 3.71QSGVEE49 pKa = 3.76RR50 pKa = 11.84PTFF53 pKa = 3.54
Molecular weight: 6.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4305 |
53 |
2191 |
615.0 |
68.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.179 ± 0.3 | 1.928 ± 0.13 |
4.785 ± 0.596 | 5.807 ± 0.076 |
4.599 ± 0.455 | 6.179 ± 0.318 |
2.23 ± 0.312 | 5.203 ± 0.399 |
5.947 ± 0.59 | 9.895 ± 0.337 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.152 | 4.019 ± 0.259 |
5.134 ± 0.402 | 3.089 ± 0.23 |
5.645 ± 0.265 | 8.548 ± 0.514 |
6.039 ± 0.41 | 8.293 ± 0.726 |
1.092 ± 0.171 | 3.415 ± 0.246 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
