
Polaribacter glomeratus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
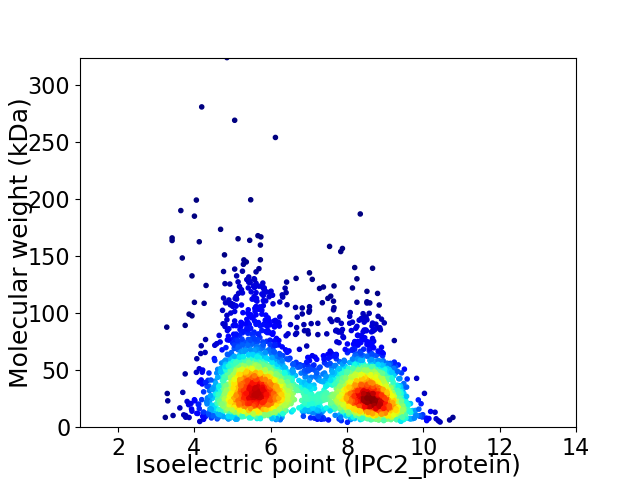
Virtual 2D-PAGE plot for 3368 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7WFZ6|A0A2S7WFZ6_9FLAO General secretion pathway protein GspG OS=Polaribacter glomeratus OX=102 GN=BTO16_11580 PE=4 SV=1
MM1 pKa = 7.32NKK3 pKa = 10.21KK4 pKa = 8.05ITLVFVSFLFAISTTFGNNDD24 pKa = 2.75NFFFEE29 pKa = 4.62NDD31 pKa = 2.57IFTVVKK37 pKa = 10.81NDD39 pKa = 3.39FKK41 pKa = 11.63SNQKK45 pKa = 8.35TVSDD49 pKa = 4.67FAAVTTTSPIISGFSSLEE67 pKa = 3.69QYY69 pKa = 11.15NNSSYY74 pKa = 10.75YY75 pKa = 10.06ISDD78 pKa = 3.54IAYY81 pKa = 7.69TWDD84 pKa = 3.49DD85 pKa = 4.68AIIATNAVGGHH96 pKa = 6.47LAVITTTAEE105 pKa = 3.59NDD107 pKa = 3.66HH108 pKa = 6.94IFAQMQAQGNQDD120 pKa = 3.38NLLIGAFEE128 pKa = 3.97SSEE131 pKa = 4.95GIFEE135 pKa = 4.46WVNGEE140 pKa = 4.38PYY142 pKa = 10.12TNNIGGAVDD151 pKa = 4.8NIPAGLNSFIVRR163 pKa = 11.84NDD165 pKa = 2.9NGGWMDD171 pKa = 3.37YY172 pKa = 10.05FGPGSSVTIKK182 pKa = 10.37IIMEE186 pKa = 4.02VPIPDD191 pKa = 3.99TIKK194 pKa = 10.75PVITLSGANPQIIEE208 pKa = 3.91VGTNYY213 pKa = 10.31TEE215 pKa = 4.74LGATATDD222 pKa = 3.68NVDD225 pKa = 3.22AANTLLVVSTVNNQDD240 pKa = 3.15TGVVGSFTVDD250 pKa = 3.28YY251 pKa = 9.35TVSDD255 pKa = 3.95VATNAAIIVTRR266 pKa = 11.84TVNVVDD272 pKa = 3.5VTKK275 pKa = 10.46PVITLAGSNPQTIEE289 pKa = 3.89VFSVYY294 pKa = 10.85DD295 pKa = 3.97DD296 pKa = 3.61LTGAIATDD304 pKa = 3.51NYY306 pKa = 10.55DD307 pKa = 4.05DD308 pKa = 4.94NVALTANIQVISNVNSSLVGTYY330 pKa = 7.4TATYY334 pKa = 9.7NVVDD338 pKa = 4.2SEE340 pKa = 5.23SNVADD345 pKa = 4.92EE346 pKa = 4.21ITKK349 pKa = 9.02TVHH352 pKa = 6.33VVDD355 pKa = 3.88TTLPEE360 pKa = 3.94IFLVGLNPQIVEE372 pKa = 3.63IGKK375 pKa = 10.4GYY377 pKa = 10.89NEE379 pKa = 4.85LGATATDD386 pKa = 3.86NYY388 pKa = 10.98DD389 pKa = 3.62PDD391 pKa = 3.72SALISNITNTSNVNTATAGSYY412 pKa = 9.72LVSYY416 pKa = 10.73NVTDD420 pKa = 4.18ANSNTAITVEE430 pKa = 4.18RR431 pKa = 11.84IVIVKK436 pKa = 10.5SNN438 pKa = 3.03
MM1 pKa = 7.32NKK3 pKa = 10.21KK4 pKa = 8.05ITLVFVSFLFAISTTFGNNDD24 pKa = 2.75NFFFEE29 pKa = 4.62NDD31 pKa = 2.57IFTVVKK37 pKa = 10.81NDD39 pKa = 3.39FKK41 pKa = 11.63SNQKK45 pKa = 8.35TVSDD49 pKa = 4.67FAAVTTTSPIISGFSSLEE67 pKa = 3.69QYY69 pKa = 11.15NNSSYY74 pKa = 10.75YY75 pKa = 10.06ISDD78 pKa = 3.54IAYY81 pKa = 7.69TWDD84 pKa = 3.49DD85 pKa = 4.68AIIATNAVGGHH96 pKa = 6.47LAVITTTAEE105 pKa = 3.59NDD107 pKa = 3.66HH108 pKa = 6.94IFAQMQAQGNQDD120 pKa = 3.38NLLIGAFEE128 pKa = 3.97SSEE131 pKa = 4.95GIFEE135 pKa = 4.46WVNGEE140 pKa = 4.38PYY142 pKa = 10.12TNNIGGAVDD151 pKa = 4.8NIPAGLNSFIVRR163 pKa = 11.84NDD165 pKa = 2.9NGGWMDD171 pKa = 3.37YY172 pKa = 10.05FGPGSSVTIKK182 pKa = 10.37IIMEE186 pKa = 4.02VPIPDD191 pKa = 3.99TIKK194 pKa = 10.75PVITLSGANPQIIEE208 pKa = 3.91VGTNYY213 pKa = 10.31TEE215 pKa = 4.74LGATATDD222 pKa = 3.68NVDD225 pKa = 3.22AANTLLVVSTVNNQDD240 pKa = 3.15TGVVGSFTVDD250 pKa = 3.28YY251 pKa = 9.35TVSDD255 pKa = 3.95VATNAAIIVTRR266 pKa = 11.84TVNVVDD272 pKa = 3.5VTKK275 pKa = 10.46PVITLAGSNPQTIEE289 pKa = 3.89VFSVYY294 pKa = 10.85DD295 pKa = 3.97DD296 pKa = 3.61LTGAIATDD304 pKa = 3.51NYY306 pKa = 10.55DD307 pKa = 4.05DD308 pKa = 4.94NVALTANIQVISNVNSSLVGTYY330 pKa = 7.4TATYY334 pKa = 9.7NVVDD338 pKa = 4.2SEE340 pKa = 5.23SNVADD345 pKa = 4.92EE346 pKa = 4.21ITKK349 pKa = 9.02TVHH352 pKa = 6.33VVDD355 pKa = 3.88TTLPEE360 pKa = 3.94IFLVGLNPQIVEE372 pKa = 3.63IGKK375 pKa = 10.4GYY377 pKa = 10.89NEE379 pKa = 4.85LGATATDD386 pKa = 3.86NYY388 pKa = 10.98DD389 pKa = 3.62PDD391 pKa = 3.72SALISNITNTSNVNTATAGSYY412 pKa = 9.72LVSYY416 pKa = 10.73NVTDD420 pKa = 4.18ANSNTAITVEE430 pKa = 4.18RR431 pKa = 11.84IVIVKK436 pKa = 10.5SNN438 pKa = 3.03
Molecular weight: 46.73 kDa
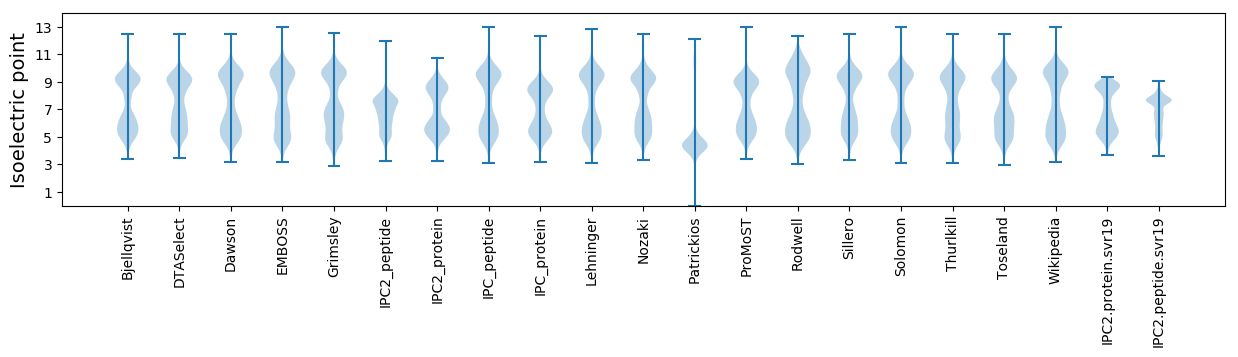
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7WK21|A0A2S7WK21_9FLAO Sulfurase OS=Polaribacter glomeratus OX=102 GN=BTO16_15980 PE=4 SV=1
MM1 pKa = 6.99QRR3 pKa = 11.84VLQRR7 pKa = 11.84VLQRR11 pKa = 11.84VLQRR15 pKa = 11.84VLQVVLLATKK25 pKa = 9.61PQGLSKK31 pKa = 11.03LIVTNIAILGIVTLLQIVLLVIKK54 pKa = 10.74LMGLNNLITSNTAIQASLSKK74 pKa = 10.78SSFMM78 pKa = 5.47
MM1 pKa = 6.99QRR3 pKa = 11.84VLQRR7 pKa = 11.84VLQRR11 pKa = 11.84VLQRR15 pKa = 11.84VLQVVLLATKK25 pKa = 9.61PQGLSKK31 pKa = 11.03LIVTNIAILGIVTLLQIVLLVIKK54 pKa = 10.74LMGLNNLITSNTAIQASLSKK74 pKa = 10.78SSFMM78 pKa = 5.47
Molecular weight: 8.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1169193 |
38 |
2942 |
347.1 |
39.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.169 ± 0.037 | 0.691 ± 0.013 |
5.347 ± 0.036 | 6.471 ± 0.042 |
5.588 ± 0.036 | 6.033 ± 0.044 |
1.602 ± 0.018 | 8.54 ± 0.04 |
8.588 ± 0.055 | 9.241 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.02 | 6.715 ± 0.047 |
3.041 ± 0.021 | 3.234 ± 0.02 |
3.07 ± 0.022 | 6.647 ± 0.033 |
6.032 ± 0.042 | 6.027 ± 0.034 |
1.009 ± 0.015 | 3.939 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
