
Hyperthermophilic Archaeal Virus 2
Taxonomy: Viruses; unclassified archaeal viruses
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
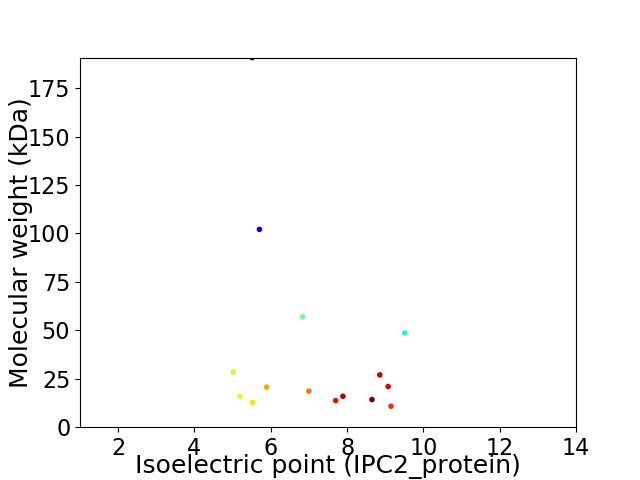
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9CGC1|D9CGC1_9VIRU Uncharacterized protein OS=Hyperthermophilic Archaeal Virus 2 OX=762906 GN=HAV2_gp12 PE=4 SV=1
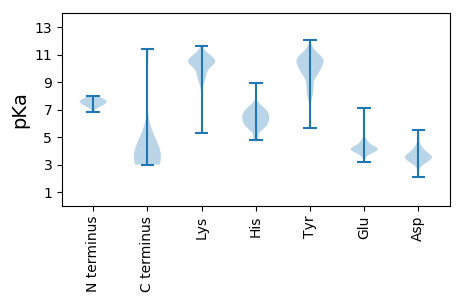
MM1 pKa = 7.99RR2 pKa = 11.84KK3 pKa = 8.83PVLLVLAVLVAGLASATLSSGQAAGVEE30 pKa = 4.53FNSVSIIVPGEE41 pKa = 3.79LLGSFQTYY49 pKa = 9.87LIPLNVSEE57 pKa = 4.71TGMLSTDD64 pKa = 3.2NVYY67 pKa = 10.93VIAQGVPRR75 pKa = 11.84FSYY78 pKa = 10.51LFSDD82 pKa = 4.4YY83 pKa = 10.39PPLVAVFEE91 pKa = 4.33RR92 pKa = 11.84SAYY95 pKa = 8.45NAVYY99 pKa = 9.12EE100 pKa = 4.15VWYY103 pKa = 10.71GGGNPYY109 pKa = 10.71SSFISVPGTGSSLWLVFDD127 pKa = 5.03DD128 pKa = 5.18FDD130 pKa = 4.95YY131 pKa = 10.83STGFWLVKK139 pKa = 9.84NASITGSRR147 pKa = 11.84VYY149 pKa = 10.72VDD151 pKa = 3.16PGGYY155 pKa = 9.54LALSAAYY162 pKa = 9.79SLRR165 pKa = 11.84TAHH168 pKa = 5.87VWLVHH173 pKa = 4.78GRR175 pKa = 11.84KK176 pKa = 9.53ALVISLQQEE185 pKa = 4.15YY186 pKa = 10.69SGFIALVFTTGNFTDD201 pKa = 3.63VSLVQNGSDD210 pKa = 3.4VFFIDD215 pKa = 4.2SLGNCLYY222 pKa = 10.94YY223 pKa = 10.91SLLHH227 pKa = 7.0LDD229 pKa = 3.52RR230 pKa = 11.84QAGIMIVVVNPAGNTVIYY248 pKa = 9.5MLYY251 pKa = 10.77GGLNKK256 pKa = 10.49CLDD259 pKa = 3.81YY260 pKa = 10.89RR261 pKa = 11.84VSS263 pKa = 3.2
MM1 pKa = 7.99RR2 pKa = 11.84KK3 pKa = 8.83PVLLVLAVLVAGLASATLSSGQAAGVEE30 pKa = 4.53FNSVSIIVPGEE41 pKa = 3.79LLGSFQTYY49 pKa = 9.87LIPLNVSEE57 pKa = 4.71TGMLSTDD64 pKa = 3.2NVYY67 pKa = 10.93VIAQGVPRR75 pKa = 11.84FSYY78 pKa = 10.51LFSDD82 pKa = 4.4YY83 pKa = 10.39PPLVAVFEE91 pKa = 4.33RR92 pKa = 11.84SAYY95 pKa = 8.45NAVYY99 pKa = 9.12EE100 pKa = 4.15VWYY103 pKa = 10.71GGGNPYY109 pKa = 10.71SSFISVPGTGSSLWLVFDD127 pKa = 5.03DD128 pKa = 5.18FDD130 pKa = 4.95YY131 pKa = 10.83STGFWLVKK139 pKa = 9.84NASITGSRR147 pKa = 11.84VYY149 pKa = 10.72VDD151 pKa = 3.16PGGYY155 pKa = 9.54LALSAAYY162 pKa = 9.79SLRR165 pKa = 11.84TAHH168 pKa = 5.87VWLVHH173 pKa = 4.78GRR175 pKa = 11.84KK176 pKa = 9.53ALVISLQQEE185 pKa = 4.15YY186 pKa = 10.69SGFIALVFTTGNFTDD201 pKa = 3.63VSLVQNGSDD210 pKa = 3.4VFFIDD215 pKa = 4.2SLGNCLYY222 pKa = 10.94YY223 pKa = 10.91SLLHH227 pKa = 7.0LDD229 pKa = 3.52RR230 pKa = 11.84QAGIMIVVVNPAGNTVIYY248 pKa = 9.5MLYY251 pKa = 10.77GGLNKK256 pKa = 10.49CLDD259 pKa = 3.81YY260 pKa = 10.89RR261 pKa = 11.84VSS263 pKa = 3.2
Molecular weight: 28.52 kDa
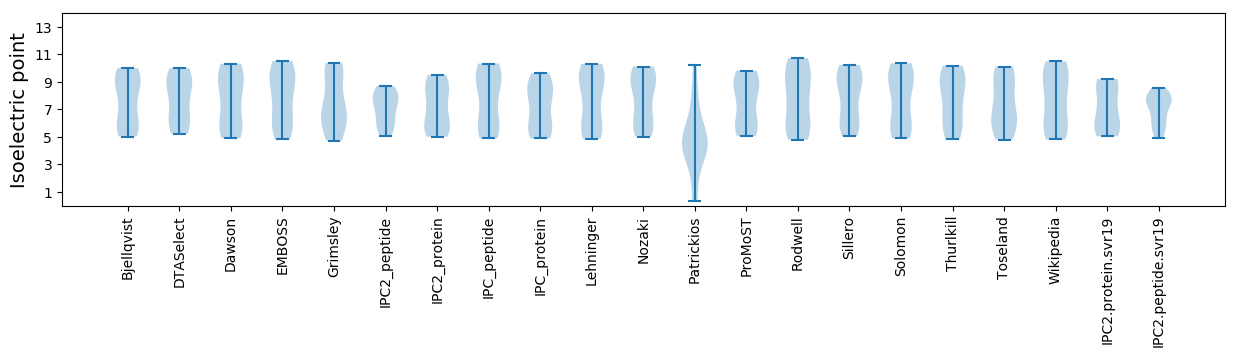
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9CGC3|D9CGC3_9VIRU Uncharacterized protein OS=Hyperthermophilic Archaeal Virus 2 OX=762906 GN=HAV2_gp14 PE=4 SV=1
MM1 pKa = 7.5LACLAYY7 pKa = 10.48SSGVSGGVASEE18 pKa = 3.95AFIIIILPSSRR29 pKa = 11.84VTRR32 pKa = 11.84SLPLITPVLAFKK44 pKa = 10.45EE45 pKa = 4.44YY46 pKa = 10.48TSLPLYY52 pKa = 10.66LSIHH56 pKa = 5.75GFFVLVLNMCMSLEE70 pKa = 3.85QLIFLTVTKK79 pKa = 10.71GMVFVNGFTGGQLHH93 pKa = 6.47EE94 pKa = 4.02RR95 pKa = 11.84FIRR98 pKa = 11.84NKK100 pKa = 9.65VFDD103 pKa = 5.48HH104 pKa = 6.34IPPRR108 pKa = 11.84AVQSRR113 pKa = 11.84LDD115 pKa = 3.37IPEE118 pKa = 4.12SFKK121 pKa = 11.23KK122 pKa = 10.46PLVALPKK129 pKa = 10.29VYY131 pKa = 10.59LNEE134 pKa = 4.03HH135 pKa = 6.15RR136 pKa = 11.84HH137 pKa = 4.94LHH139 pKa = 5.88PLFPSPLTSAPSGLTLSIRR158 pKa = 11.84SRR160 pKa = 11.84ILRR163 pKa = 11.84NSLDD167 pKa = 3.58INTSTASAIILNAWAEE183 pKa = 4.16MGFRR187 pKa = 11.84PCSS190 pKa = 3.31
MM1 pKa = 7.5LACLAYY7 pKa = 10.48SSGVSGGVASEE18 pKa = 3.95AFIIIILPSSRR29 pKa = 11.84VTRR32 pKa = 11.84SLPLITPVLAFKK44 pKa = 10.45EE45 pKa = 4.44YY46 pKa = 10.48TSLPLYY52 pKa = 10.66LSIHH56 pKa = 5.75GFFVLVLNMCMSLEE70 pKa = 3.85QLIFLTVTKK79 pKa = 10.71GMVFVNGFTGGQLHH93 pKa = 6.47EE94 pKa = 4.02RR95 pKa = 11.84FIRR98 pKa = 11.84NKK100 pKa = 9.65VFDD103 pKa = 5.48HH104 pKa = 6.34IPPRR108 pKa = 11.84AVQSRR113 pKa = 11.84LDD115 pKa = 3.37IPEE118 pKa = 4.12SFKK121 pKa = 11.23KK122 pKa = 10.46PLVALPKK129 pKa = 10.29VYY131 pKa = 10.59LNEE134 pKa = 4.03HH135 pKa = 6.15RR136 pKa = 11.84HH137 pKa = 4.94LHH139 pKa = 5.88PLFPSPLTSAPSGLTLSIRR158 pKa = 11.84SRR160 pKa = 11.84ILRR163 pKa = 11.84NSLDD167 pKa = 3.58INTSTASAIILNAWAEE183 pKa = 4.16MGFRR187 pKa = 11.84PCSS190 pKa = 3.31
Molecular weight: 20.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5384 |
93 |
1767 |
358.9 |
39.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.689 ± 0.244 | 0.743 ± 0.174 |
4.123 ± 0.234 | 5.665 ± 1.276 |
3.529 ± 0.258 | 7.207 ± 0.544 |
1.449 ± 0.273 | 5.609 ± 0.466 |
4.941 ± 1.122 | 10.048 ± 0.512 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.746 ± 0.264 | 3.845 ± 0.528 |
5.126 ± 0.209 | 2.025 ± 0.172 |
5.479 ± 0.631 | 8.042 ± 0.901 |
5.981 ± 0.967 | 9.733 ± 0.667 |
1.597 ± 0.124 | 5.405 ± 0.747 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
