
Slackia piriformis YIT 12062
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Slackia; Slackia piriformis
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
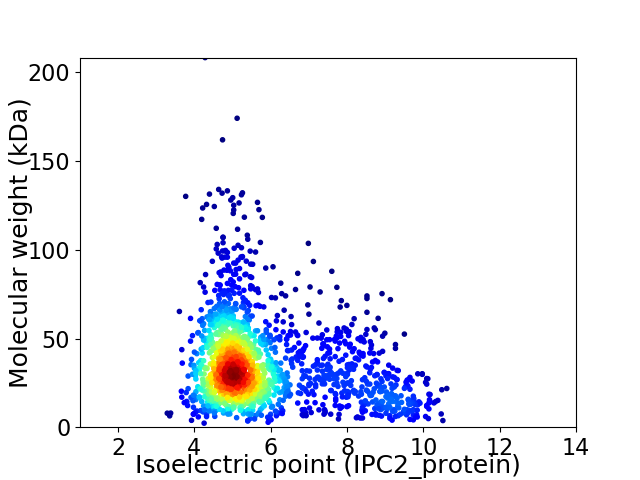
Virtual 2D-PAGE plot for 1794 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0YIV2|K0YIV2_9ACTN HD domain-containing protein OS=Slackia piriformis YIT 12062 OX=742818 GN=HMPREF9451_01007 PE=4 SV=1
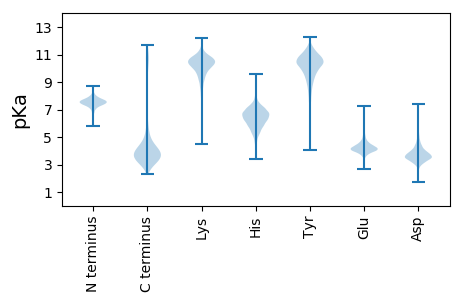
MM1 pKa = 7.54EE2 pKa = 5.0EE3 pKa = 4.39KK4 pKa = 10.22KK5 pKa = 11.04GPAAPQPAPDD15 pKa = 4.25NQPKK19 pKa = 10.31SDD21 pKa = 3.46ISAEE25 pKa = 4.07PAAPAAPVEE34 pKa = 4.63PPAPTAPAAAPAEE47 pKa = 4.19PAAPAEE53 pKa = 4.26PAAPAEE59 pKa = 4.2PAVPAEE65 pKa = 4.18PAAPAEE71 pKa = 4.5PAAPMAPVAPTAPAAPQAAPASAPQVEE98 pKa = 5.16GTAQMPQTPHH108 pKa = 6.22QPVSAPTQPFDD119 pKa = 4.47SAQQPAQPVPGPAQAPYY136 pKa = 10.34GAYY139 pKa = 9.33PPPPAPNYY147 pKa = 8.93SAPSSGKK154 pKa = 8.5ATGALICGILAIVFSFIPIVGIILGIVAIVLASKK188 pKa = 10.26AVKK191 pKa = 9.79FAGKK195 pKa = 9.76SGKK198 pKa = 6.23TTGGKK203 pKa = 8.74VCGIIGIVFSILWAIFSFVLTMGIIGGINAYY234 pKa = 9.84EE235 pKa = 4.1SDD237 pKa = 4.55YY238 pKa = 11.33VPSADD243 pKa = 3.93VPTTSSPLAEE253 pKa = 4.47DD254 pKa = 3.27MGAEE258 pKa = 4.06EE259 pKa = 4.4QAIADD264 pKa = 4.18LVSAEE269 pKa = 3.72LDD271 pKa = 3.32KK272 pKa = 11.72YY273 pKa = 11.35KK274 pKa = 10.96NADD277 pKa = 3.48SAALSALGAEE287 pKa = 4.74YY288 pKa = 10.6DD289 pKa = 4.56DD290 pKa = 4.95EE291 pKa = 5.65DD292 pKa = 4.68LVTIDD297 pKa = 4.37GVDD300 pKa = 3.3MSLSDD305 pKa = 5.35LGIDD309 pKa = 3.88PNEE312 pKa = 3.81VAAWATTDD320 pKa = 3.59FDD322 pKa = 4.48YY323 pKa = 11.4SIDD326 pKa = 3.64SVYY329 pKa = 10.43IDD331 pKa = 4.14SDD333 pKa = 3.8GATATVYY340 pKa = 11.03VDD342 pKa = 3.05TTEE345 pKa = 4.49RR346 pKa = 11.84DD347 pKa = 3.63FFEE350 pKa = 4.41MGSLFGDD357 pKa = 4.3LLNEE361 pKa = 4.23YY362 pKa = 9.36VSSGAADD369 pKa = 3.53AADD372 pKa = 4.62LEE374 pKa = 4.57TAKK377 pKa = 11.03AKK379 pKa = 10.7VGEE382 pKa = 4.63LYY384 pKa = 10.41AQAMAEE390 pKa = 4.16STAMSDD396 pKa = 3.25YY397 pKa = 10.72YY398 pKa = 11.46AAFDD402 pKa = 3.57IVNEE406 pKa = 4.3GGTWRR411 pKa = 11.84ILEE414 pKa = 4.46DD415 pKa = 3.62SLEE418 pKa = 4.15EE419 pKa = 3.78EE420 pKa = 4.04LDD422 pKa = 3.92YY423 pKa = 11.68VFSTFF428 pKa = 4.33
MM1 pKa = 7.54EE2 pKa = 5.0EE3 pKa = 4.39KK4 pKa = 10.22KK5 pKa = 11.04GPAAPQPAPDD15 pKa = 4.25NQPKK19 pKa = 10.31SDD21 pKa = 3.46ISAEE25 pKa = 4.07PAAPAAPVEE34 pKa = 4.63PPAPTAPAAAPAEE47 pKa = 4.19PAAPAEE53 pKa = 4.26PAAPAEE59 pKa = 4.2PAVPAEE65 pKa = 4.18PAAPAEE71 pKa = 4.5PAAPMAPVAPTAPAAPQAAPASAPQVEE98 pKa = 5.16GTAQMPQTPHH108 pKa = 6.22QPVSAPTQPFDD119 pKa = 4.47SAQQPAQPVPGPAQAPYY136 pKa = 10.34GAYY139 pKa = 9.33PPPPAPNYY147 pKa = 8.93SAPSSGKK154 pKa = 8.5ATGALICGILAIVFSFIPIVGIILGIVAIVLASKK188 pKa = 10.26AVKK191 pKa = 9.79FAGKK195 pKa = 9.76SGKK198 pKa = 6.23TTGGKK203 pKa = 8.74VCGIIGIVFSILWAIFSFVLTMGIIGGINAYY234 pKa = 9.84EE235 pKa = 4.1SDD237 pKa = 4.55YY238 pKa = 11.33VPSADD243 pKa = 3.93VPTTSSPLAEE253 pKa = 4.47DD254 pKa = 3.27MGAEE258 pKa = 4.06EE259 pKa = 4.4QAIADD264 pKa = 4.18LVSAEE269 pKa = 3.72LDD271 pKa = 3.32KK272 pKa = 11.72YY273 pKa = 11.35KK274 pKa = 10.96NADD277 pKa = 3.48SAALSALGAEE287 pKa = 4.74YY288 pKa = 10.6DD289 pKa = 4.56DD290 pKa = 4.95EE291 pKa = 5.65DD292 pKa = 4.68LVTIDD297 pKa = 4.37GVDD300 pKa = 3.3MSLSDD305 pKa = 5.35LGIDD309 pKa = 3.88PNEE312 pKa = 3.81VAAWATTDD320 pKa = 3.59FDD322 pKa = 4.48YY323 pKa = 11.4SIDD326 pKa = 3.64SVYY329 pKa = 10.43IDD331 pKa = 4.14SDD333 pKa = 3.8GATATVYY340 pKa = 11.03VDD342 pKa = 3.05TTEE345 pKa = 4.49RR346 pKa = 11.84DD347 pKa = 3.63FFEE350 pKa = 4.41MGSLFGDD357 pKa = 4.3LLNEE361 pKa = 4.23YY362 pKa = 9.36VSSGAADD369 pKa = 3.53AADD372 pKa = 4.62LEE374 pKa = 4.57TAKK377 pKa = 11.03AKK379 pKa = 10.7VGEE382 pKa = 4.63LYY384 pKa = 10.41AQAMAEE390 pKa = 4.16STAMSDD396 pKa = 3.25YY397 pKa = 10.72YY398 pKa = 11.46AAFDD402 pKa = 3.57IVNEE406 pKa = 4.3GGTWRR411 pKa = 11.84ILEE414 pKa = 4.46DD415 pKa = 3.62SLEE418 pKa = 4.15EE419 pKa = 3.78EE420 pKa = 4.04LDD422 pKa = 3.92YY423 pKa = 11.68VFSTFF428 pKa = 4.33
Molecular weight: 43.72 kDa
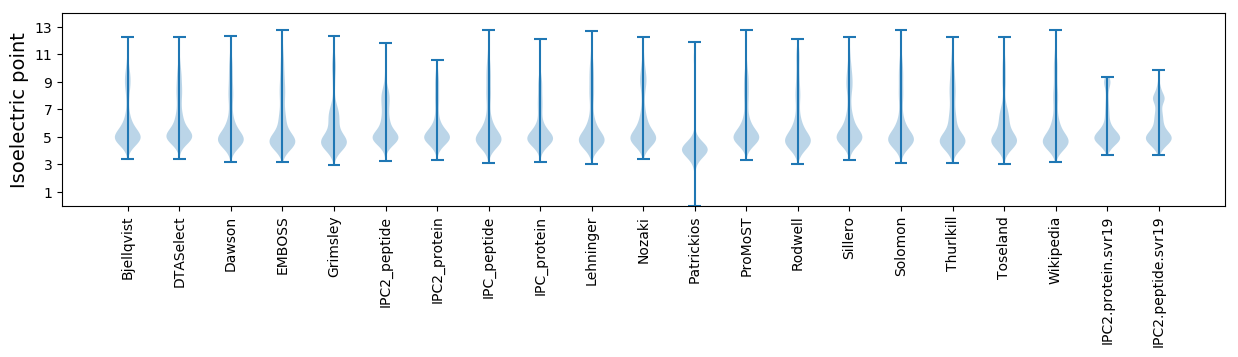
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0YL01|K0YL01_9ACTN Uncharacterized protein OS=Slackia piriformis YIT 12062 OX=742818 GN=HMPREF9451_00882 PE=4 SV=1
MM1 pKa = 7.18EE2 pKa = 5.86RR3 pKa = 11.84KK4 pKa = 9.19PVDD7 pKa = 3.48EE8 pKa = 4.45YY9 pKa = 11.62LATGPVEE16 pKa = 3.96EE17 pKa = 4.56SSFRR21 pKa = 11.84SVILFGRR28 pKa = 11.84NTASYY33 pKa = 10.74KK34 pKa = 10.3FALAKK39 pKa = 10.68ALFDD43 pKa = 4.28LAGQNRR49 pKa = 11.84DD50 pKa = 3.44SVTLNQLAVPYY61 pKa = 8.55TKK63 pKa = 10.44HH64 pKa = 6.37LVEE67 pKa = 4.8HH68 pKa = 6.83AGSNPRR74 pKa = 11.84QTTSRR79 pKa = 11.84SSRR82 pKa = 11.84FMDD85 pKa = 3.48ACVGYY90 pKa = 10.83AKK92 pKa = 10.49GSEE95 pKa = 4.03IYY97 pKa = 10.29DD98 pKa = 3.52RR99 pKa = 11.84LISAVVDD106 pKa = 3.87LGFDD110 pKa = 3.31NVLDD114 pKa = 4.18AFHH117 pKa = 6.54TVGKK121 pKa = 10.31GDD123 pKa = 3.47VPVRR127 pKa = 11.84FSKK130 pKa = 10.79RR131 pKa = 11.84IFRR134 pKa = 11.84GDD136 pKa = 3.42RR137 pKa = 11.84NVSSLPTRR145 pKa = 11.84SSNWRR150 pKa = 11.84IRR152 pKa = 11.84PRR154 pKa = 11.84RR155 pKa = 11.84PASFKK160 pKa = 10.58KK161 pKa = 10.39SNRR164 pKa = 11.84DD165 pKa = 3.3GASSRR170 pKa = 11.84PPGRR174 pKa = 11.84PASALAFFPMMEE186 pKa = 4.31RR187 pKa = 11.84PGGSCLMAVCAEE199 pKa = 3.94GTLRR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84ASRR208 pKa = 3.31
MM1 pKa = 7.18EE2 pKa = 5.86RR3 pKa = 11.84KK4 pKa = 9.19PVDD7 pKa = 3.48EE8 pKa = 4.45YY9 pKa = 11.62LATGPVEE16 pKa = 3.96EE17 pKa = 4.56SSFRR21 pKa = 11.84SVILFGRR28 pKa = 11.84NTASYY33 pKa = 10.74KK34 pKa = 10.3FALAKK39 pKa = 10.68ALFDD43 pKa = 4.28LAGQNRR49 pKa = 11.84DD50 pKa = 3.44SVTLNQLAVPYY61 pKa = 8.55TKK63 pKa = 10.44HH64 pKa = 6.37LVEE67 pKa = 4.8HH68 pKa = 6.83AGSNPRR74 pKa = 11.84QTTSRR79 pKa = 11.84SSRR82 pKa = 11.84FMDD85 pKa = 3.48ACVGYY90 pKa = 10.83AKK92 pKa = 10.49GSEE95 pKa = 4.03IYY97 pKa = 10.29DD98 pKa = 3.52RR99 pKa = 11.84LISAVVDD106 pKa = 3.87LGFDD110 pKa = 3.31NVLDD114 pKa = 4.18AFHH117 pKa = 6.54TVGKK121 pKa = 10.31GDD123 pKa = 3.47VPVRR127 pKa = 11.84FSKK130 pKa = 10.79RR131 pKa = 11.84IFRR134 pKa = 11.84GDD136 pKa = 3.42RR137 pKa = 11.84NVSSLPTRR145 pKa = 11.84SSNWRR150 pKa = 11.84IRR152 pKa = 11.84PRR154 pKa = 11.84RR155 pKa = 11.84PASFKK160 pKa = 10.58KK161 pKa = 10.39SNRR164 pKa = 11.84DD165 pKa = 3.3GASSRR170 pKa = 11.84PPGRR174 pKa = 11.84PASALAFFPMMEE186 pKa = 4.31RR187 pKa = 11.84PGGSCLMAVCAEE199 pKa = 3.94GTLRR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84ASRR208 pKa = 3.31
Molecular weight: 23.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
600490 |
22 |
1949 |
334.7 |
36.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.322 ± 0.068 | 1.74 ± 0.028 |
5.936 ± 0.048 | 6.906 ± 0.065 |
4.089 ± 0.038 | 8.03 ± 0.053 |
1.861 ± 0.024 | 5.313 ± 0.047 |
4.176 ± 0.043 | 8.928 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.024 | 2.917 ± 0.031 |
4.07 ± 0.031 | 2.78 ± 0.03 |
5.77 ± 0.069 | 6.408 ± 0.048 |
4.886 ± 0.041 | 8.139 ± 0.053 |
1.016 ± 0.021 | 2.83 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
