
Saimiriine herpesvirus 1 (strain MV-5-4-PSL) (SaHV-1) (Marmoset herpesvirus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
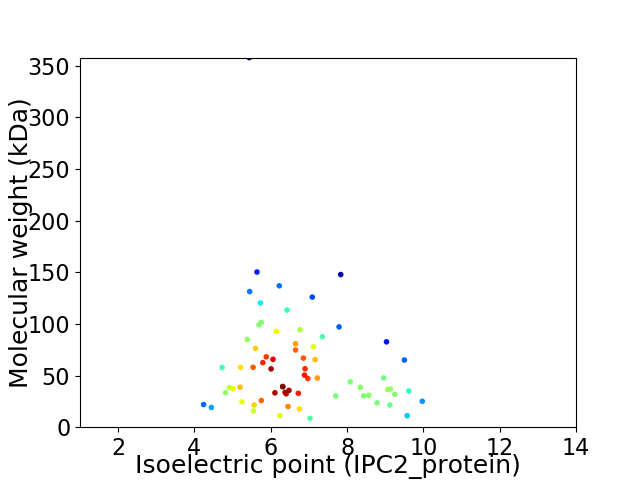
Virtual 2D-PAGE plot for 69 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2IUG7|E2IUG7_SHV1 Nuclear protein UL3 OS=Saimiriine herpesvirus 1 (strain MV-5-4-PSL) OX=10353 GN=UL03 PE=3 SV=1
MM1 pKa = 7.98DD2 pKa = 5.85PSSQLSLVAYY12 pKa = 6.88TLRR15 pKa = 11.84NARR18 pKa = 11.84GPTTWKK24 pKa = 10.71LYY26 pKa = 8.71DD27 pKa = 3.53TEE29 pKa = 5.8HH30 pKa = 6.53IVSTFDD36 pKa = 2.99SGVRR40 pKa = 11.84YY41 pKa = 9.52VATAGRR47 pKa = 11.84LRR49 pKa = 11.84QDD51 pKa = 3.64PLDD54 pKa = 4.19GGSVVLQDD62 pKa = 3.5TPGGVVVVVNCPADD76 pKa = 3.81FCAYY80 pKa = 9.83RR81 pKa = 11.84FVGRR85 pKa = 11.84TQGHH89 pKa = 5.97VLRR92 pKa = 11.84QWEE95 pKa = 4.23DD96 pKa = 2.58TGMCVYY102 pKa = 10.5PFDD105 pKa = 3.55AWTGSTHH112 pKa = 5.26TEE114 pKa = 4.12SVRR117 pKa = 11.84SASAGTAIVLWCDD130 pKa = 3.04DD131 pKa = 3.75SLYY134 pKa = 9.51ITATVCDD141 pKa = 4.67ADD143 pKa = 4.51PPADD147 pKa = 3.75SPTARR152 pKa = 11.84VDD154 pKa = 3.5EE155 pKa = 4.72TPGSLLEE162 pKa = 4.64DD163 pKa = 3.16AHH165 pKa = 6.88EE166 pKa = 4.33EE167 pKa = 3.96LLAGDD172 pKa = 3.75SEE174 pKa = 4.6ARR176 pKa = 11.84VAADD180 pKa = 4.38LLTEE184 pKa = 4.22VLDD187 pKa = 4.06QVQLGPASSFEE198 pKa = 3.97GCARR202 pKa = 11.84GAQQ205 pKa = 3.4
MM1 pKa = 7.98DD2 pKa = 5.85PSSQLSLVAYY12 pKa = 6.88TLRR15 pKa = 11.84NARR18 pKa = 11.84GPTTWKK24 pKa = 10.71LYY26 pKa = 8.71DD27 pKa = 3.53TEE29 pKa = 5.8HH30 pKa = 6.53IVSTFDD36 pKa = 2.99SGVRR40 pKa = 11.84YY41 pKa = 9.52VATAGRR47 pKa = 11.84LRR49 pKa = 11.84QDD51 pKa = 3.64PLDD54 pKa = 4.19GGSVVLQDD62 pKa = 3.5TPGGVVVVVNCPADD76 pKa = 3.81FCAYY80 pKa = 9.83RR81 pKa = 11.84FVGRR85 pKa = 11.84TQGHH89 pKa = 5.97VLRR92 pKa = 11.84QWEE95 pKa = 4.23DD96 pKa = 2.58TGMCVYY102 pKa = 10.5PFDD105 pKa = 3.55AWTGSTHH112 pKa = 5.26TEE114 pKa = 4.12SVRR117 pKa = 11.84SASAGTAIVLWCDD130 pKa = 3.04DD131 pKa = 3.75SLYY134 pKa = 9.51ITATVCDD141 pKa = 4.67ADD143 pKa = 4.51PPADD147 pKa = 3.75SPTARR152 pKa = 11.84VDD154 pKa = 3.5EE155 pKa = 4.72TPGSLLEE162 pKa = 4.64DD163 pKa = 3.16AHH165 pKa = 6.88EE166 pKa = 4.33EE167 pKa = 3.96LLAGDD172 pKa = 3.75SEE174 pKa = 4.6ARR176 pKa = 11.84VAADD180 pKa = 4.38LLTEE184 pKa = 4.22VLDD187 pKa = 4.06QVQLGPASSFEE198 pKa = 3.97GCARR202 pKa = 11.84GAQQ205 pKa = 3.4
Molecular weight: 21.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2IUG9|E2IUG9_SHV1 Envelope glycoprotein L OS=Saimiriine herpesvirus 1 (strain MV-5-4-PSL) OX=10353 GN=UL01 PE=3 SV=1
MM1 pKa = 7.39KK2 pKa = 10.32RR3 pKa = 11.84QATPDD8 pKa = 3.39SRR10 pKa = 11.84QLTLLAPQLPSPSNHH25 pKa = 6.63PPPLPEE31 pKa = 3.82SRR33 pKa = 11.84RR34 pKa = 11.84GAEE37 pKa = 3.98DD38 pKa = 3.58PKK40 pKa = 10.85TEE42 pKa = 4.21GSSAPDD48 pKa = 3.49VIEE51 pKa = 4.58PPLAAVAKK59 pKa = 9.86RR60 pKa = 11.84PRR62 pKa = 11.84RR63 pKa = 11.84PRR65 pKa = 11.84GCPLGVQFGGSPSAADD81 pKa = 3.1AVEE84 pKa = 4.65RR85 pKa = 11.84EE86 pKa = 4.35TASLFSPSSGVSDD99 pKa = 3.64RR100 pKa = 11.84TLDD103 pKa = 3.08WRR105 pKa = 11.84RR106 pKa = 11.84ARR108 pKa = 11.84DD109 pKa = 3.35VFGIGEE115 pKa = 4.17PWRR118 pKa = 11.84DD119 pKa = 3.53VIEE122 pKa = 4.76PEE124 pKa = 3.88LAAAATSGLLSEE136 pKa = 4.38YY137 pKa = 10.13FRR139 pKa = 11.84RR140 pKa = 11.84CRR142 pKa = 11.84TEE144 pKa = 3.88EE145 pKa = 3.85VLPPRR150 pKa = 11.84GDD152 pKa = 3.34VFSWTRR158 pKa = 11.84SCAPDD163 pKa = 3.55DD164 pKa = 3.67VRR166 pKa = 11.84VVIIGQDD173 pKa = 3.65PYY175 pKa = 10.43HH176 pKa = 6.66QPGQAHH182 pKa = 6.65GLAFSVRR189 pKa = 11.84PGVPVPPSLRR199 pKa = 11.84NIFTAVRR206 pKa = 11.84SCYY209 pKa = 10.14PEE211 pKa = 3.88ATPAAGHH218 pKa = 6.2GCLEE222 pKa = 3.62KK223 pKa = 10.01WARR226 pKa = 11.84QGVLLLNTTLTVRR239 pKa = 11.84RR240 pKa = 11.84GAAGSHH246 pKa = 6.36SGLGWGRR253 pKa = 11.84FVAGVLRR260 pKa = 11.84RR261 pKa = 11.84LAQRR265 pKa = 11.84RR266 pKa = 11.84DD267 pKa = 3.28GLVFMLWGQHH277 pKa = 4.63AQGAIKK283 pKa = 10.28PDD285 pKa = 3.77TQRR288 pKa = 11.84HH289 pKa = 4.23LVLRR293 pKa = 11.84FAHH296 pKa = 6.86PSPLSQTPFGTCRR309 pKa = 11.84HH310 pKa = 5.8FLLANQYY317 pKa = 10.71LEE319 pKa = 4.95ARR321 pKa = 11.84GLPPIDD327 pKa = 3.68WSLGPAPSLARR338 pKa = 11.84AKK340 pKa = 10.59
MM1 pKa = 7.39KK2 pKa = 10.32RR3 pKa = 11.84QATPDD8 pKa = 3.39SRR10 pKa = 11.84QLTLLAPQLPSPSNHH25 pKa = 6.63PPPLPEE31 pKa = 3.82SRR33 pKa = 11.84RR34 pKa = 11.84GAEE37 pKa = 3.98DD38 pKa = 3.58PKK40 pKa = 10.85TEE42 pKa = 4.21GSSAPDD48 pKa = 3.49VIEE51 pKa = 4.58PPLAAVAKK59 pKa = 9.86RR60 pKa = 11.84PRR62 pKa = 11.84RR63 pKa = 11.84PRR65 pKa = 11.84GCPLGVQFGGSPSAADD81 pKa = 3.1AVEE84 pKa = 4.65RR85 pKa = 11.84EE86 pKa = 4.35TASLFSPSSGVSDD99 pKa = 3.64RR100 pKa = 11.84TLDD103 pKa = 3.08WRR105 pKa = 11.84RR106 pKa = 11.84ARR108 pKa = 11.84DD109 pKa = 3.35VFGIGEE115 pKa = 4.17PWRR118 pKa = 11.84DD119 pKa = 3.53VIEE122 pKa = 4.76PEE124 pKa = 3.88LAAAATSGLLSEE136 pKa = 4.38YY137 pKa = 10.13FRR139 pKa = 11.84RR140 pKa = 11.84CRR142 pKa = 11.84TEE144 pKa = 3.88EE145 pKa = 3.85VLPPRR150 pKa = 11.84GDD152 pKa = 3.34VFSWTRR158 pKa = 11.84SCAPDD163 pKa = 3.55DD164 pKa = 3.67VRR166 pKa = 11.84VVIIGQDD173 pKa = 3.65PYY175 pKa = 10.43HH176 pKa = 6.66QPGQAHH182 pKa = 6.65GLAFSVRR189 pKa = 11.84PGVPVPPSLRR199 pKa = 11.84NIFTAVRR206 pKa = 11.84SCYY209 pKa = 10.14PEE211 pKa = 3.88ATPAAGHH218 pKa = 6.2GCLEE222 pKa = 3.62KK223 pKa = 10.01WARR226 pKa = 11.84QGVLLLNTTLTVRR239 pKa = 11.84RR240 pKa = 11.84GAAGSHH246 pKa = 6.36SGLGWGRR253 pKa = 11.84FVAGVLRR260 pKa = 11.84RR261 pKa = 11.84LAQRR265 pKa = 11.84RR266 pKa = 11.84DD267 pKa = 3.28GLVFMLWGQHH277 pKa = 4.63AQGAIKK283 pKa = 10.28PDD285 pKa = 3.77TQRR288 pKa = 11.84HH289 pKa = 4.23LVLRR293 pKa = 11.84FAHH296 pKa = 6.86PSPLSQTPFGTCRR309 pKa = 11.84HH310 pKa = 5.8FLLANQYY317 pKa = 10.71LEE319 pKa = 4.95ARR321 pKa = 11.84GLPPIDD327 pKa = 3.68WSLGPAPSLARR338 pKa = 11.84AKK340 pKa = 10.59
Molecular weight: 36.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
38775 |
76 |
3390 |
562.0 |
60.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.025 ± 0.557 | 1.782 ± 0.115 |
5.638 ± 0.175 | 5.594 ± 0.155 |
3.317 ± 0.187 | 7.319 ± 0.283 |
2.218 ± 0.107 | 2.507 ± 0.173 |
1.666 ± 0.118 | 9.439 ± 0.254 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.586 ± 0.097 | 1.95 ± 0.163 |
7.892 ± 0.445 | 2.545 ± 0.143 |
9.32 ± 0.28 | 7.51 ± 0.227 |
5.207 ± 0.196 | 6.894 ± 0.241 |
1.13 ± 0.065 | 2.463 ± 0.154 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
