
Human papillomavirus type 156
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 18
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
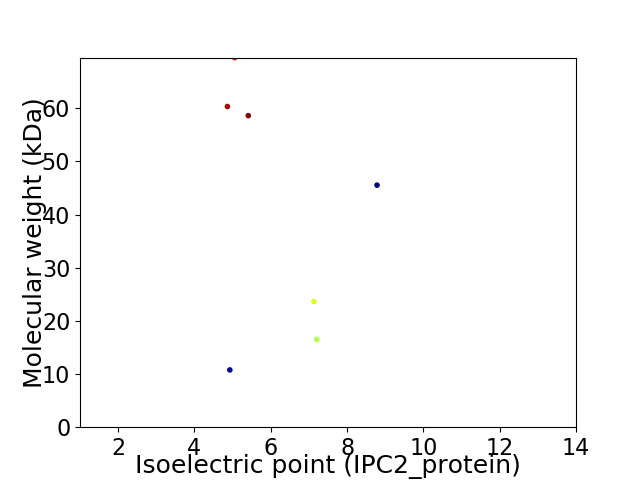
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7QNV2|K7QNV2_9PAPI Major capsid protein L1 OS=Human papillomavirus type 156 OX=1248396 GN=L1 PE=3 SV=1
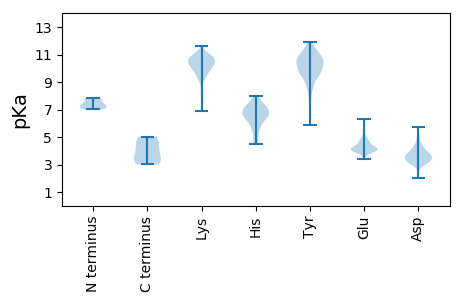
MM1 pKa = 7.48IGNRR5 pKa = 11.84PDD7 pKa = 3.15IKK9 pKa = 10.88DD10 pKa = 3.32IEE12 pKa = 5.23LDD14 pKa = 3.37LHH16 pKa = 7.05ALVLPANLLSEE27 pKa = 4.67EE28 pKa = 4.24SLSPDD33 pKa = 3.08SEE35 pKa = 4.39AEE37 pKa = 3.99EE38 pKa = 4.52EE39 pKa = 4.71EE40 pKa = 4.05EE41 pKa = 4.0HH42 pKa = 6.65SPYY45 pKa = 10.65RR46 pKa = 11.84VDD48 pKa = 3.57TCCKK52 pKa = 9.36SCGTGVRR59 pKa = 11.84LCVFATQLAIRR70 pKa = 11.84TLQQLLTQEE79 pKa = 4.66LSLVCPGCSRR89 pKa = 11.84NLFHH93 pKa = 7.8HH94 pKa = 6.52GRR96 pKa = 11.84LL97 pKa = 3.68
MM1 pKa = 7.48IGNRR5 pKa = 11.84PDD7 pKa = 3.15IKK9 pKa = 10.88DD10 pKa = 3.32IEE12 pKa = 5.23LDD14 pKa = 3.37LHH16 pKa = 7.05ALVLPANLLSEE27 pKa = 4.67EE28 pKa = 4.24SLSPDD33 pKa = 3.08SEE35 pKa = 4.39AEE37 pKa = 3.99EE38 pKa = 4.52EE39 pKa = 4.71EE40 pKa = 4.05EE41 pKa = 4.0HH42 pKa = 6.65SPYY45 pKa = 10.65RR46 pKa = 11.84VDD48 pKa = 3.57TCCKK52 pKa = 9.36SCGTGVRR59 pKa = 11.84LCVFATQLAIRR70 pKa = 11.84TLQQLLTQEE79 pKa = 4.66LSLVCPGCSRR89 pKa = 11.84NLFHH93 pKa = 7.8HH94 pKa = 6.52GRR96 pKa = 11.84LL97 pKa = 3.68
Molecular weight: 10.78 kDa
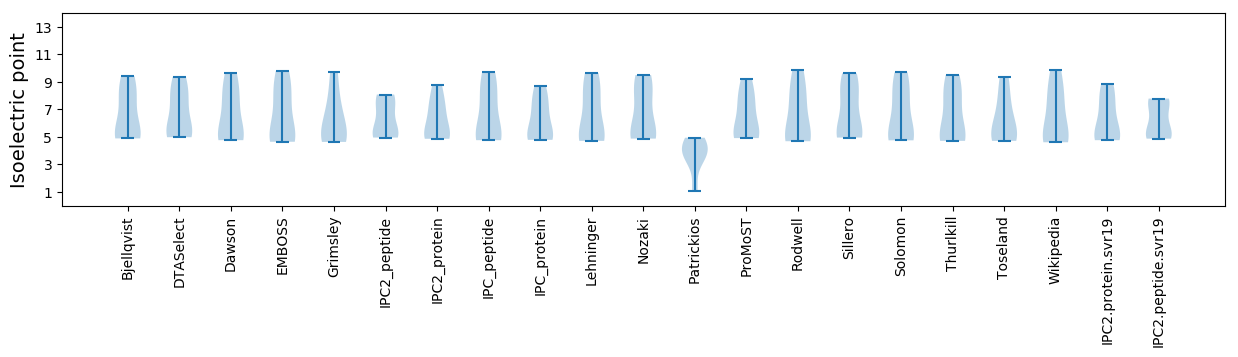
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7QR99|K7QR99_9PAPI Protein E6 OS=Human papillomavirus type 156 OX=1248396 GN=E6 PE=3 SV=1
MM1 pKa = 7.02NQADD5 pKa = 3.85LTVRR9 pKa = 11.84FDD11 pKa = 3.76ALQEE15 pKa = 4.01RR16 pKa = 11.84LMTLYY21 pKa = 10.92EE22 pKa = 4.27EE23 pKa = 4.61GRR25 pKa = 11.84KK26 pKa = 8.15TIDD29 pKa = 3.05AQIEE33 pKa = 4.05YY34 pKa = 8.63WQLIRR39 pKa = 11.84KK40 pKa = 8.64EE41 pKa = 4.36SVLMYY46 pKa = 9.58YY47 pKa = 10.26GRR49 pKa = 11.84KK50 pKa = 9.56EE51 pKa = 3.82GFKK54 pKa = 11.0NFGLQPLPALTVSEE68 pKa = 4.61YY69 pKa = 10.14KK70 pKa = 10.62AKK72 pKa = 10.22EE73 pKa = 4.1AIRR76 pKa = 11.84QVLLLKK82 pKa = 10.3SLKK85 pKa = 10.62NSVFGNEE92 pKa = 3.86EE93 pKa = 3.42WTLPEE98 pKa = 3.82TSAEE102 pKa = 4.14LTYY105 pKa = 9.67TAPRR109 pKa = 11.84NTFKK113 pKa = 10.88KK114 pKa = 10.38DD115 pKa = 3.05PYY117 pKa = 10.42TVDD120 pKa = 2.01VWFDD124 pKa = 3.8HH125 pKa = 6.84NPDD128 pKa = 3.01NSFPYY133 pKa = 10.03TNWNKK138 pKa = 9.97IYY140 pKa = 10.35FQGDD144 pKa = 2.95HH145 pKa = 7.21DD146 pKa = 4.0NWHH149 pKa = 6.32RR150 pKa = 11.84AAGKK154 pKa = 10.14VDD156 pKa = 3.24INGLYY161 pKa = 10.5FEE163 pKa = 5.55DD164 pKa = 4.09ANGDD168 pKa = 2.96KK169 pKa = 10.97SYY171 pKa = 11.0FVVFASDD178 pKa = 3.11AQRR181 pKa = 11.84YY182 pKa = 5.84GTTGEE187 pKa = 4.01WTVHH191 pKa = 5.35YY192 pKa = 10.09KK193 pKa = 10.71NEE195 pKa = 4.7TISSTSLSSQRR206 pKa = 11.84SLSTISSQGSVSSSRR221 pKa = 11.84DD222 pKa = 3.31SIPAKK227 pKa = 10.05KK228 pKa = 9.88GQSTRR233 pKa = 11.84RR234 pKa = 11.84HH235 pKa = 4.56QSEE238 pKa = 4.07EE239 pKa = 4.32GIADD243 pKa = 3.7SSSSSTSSSPSAVRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84PQQGEE267 pKa = 3.94HH268 pKa = 5.46TSRR271 pKa = 11.84GGGTRR276 pKa = 11.84AKK278 pKa = 9.93RR279 pKa = 11.84QRR281 pKa = 11.84TEE283 pKa = 3.98EE284 pKa = 4.03VSTTVSPDD292 pKa = 2.85EE293 pKa = 4.15VGRR296 pKa = 11.84GNTSVPRR303 pKa = 11.84TNLTRR308 pKa = 11.84LEE310 pKa = 4.12RR311 pKa = 11.84LAKK314 pKa = 9.69EE315 pKa = 4.22ARR317 pKa = 11.84DD318 pKa = 3.72PPIVLIKK325 pKa = 10.68GRR327 pKa = 11.84SNPLKK332 pKa = 9.17CWRR335 pKa = 11.84YY336 pKa = 9.56RR337 pKa = 11.84CEE339 pKa = 4.1KK340 pKa = 10.72HH341 pKa = 6.68CDD343 pKa = 3.11LYY345 pKa = 11.68DD346 pKa = 4.97RR347 pKa = 11.84ISTVFKK353 pKa = 10.02WVNSTNDD360 pKa = 3.46CNMHH364 pKa = 6.72RR365 pKa = 11.84LLISFKK371 pKa = 10.42NQNQRR376 pKa = 11.84SMFLKK381 pKa = 10.44FVNLPKK387 pKa = 10.88GCTFSLGSLDD397 pKa = 4.62SII399 pKa = 5.04
MM1 pKa = 7.02NQADD5 pKa = 3.85LTVRR9 pKa = 11.84FDD11 pKa = 3.76ALQEE15 pKa = 4.01RR16 pKa = 11.84LMTLYY21 pKa = 10.92EE22 pKa = 4.27EE23 pKa = 4.61GRR25 pKa = 11.84KK26 pKa = 8.15TIDD29 pKa = 3.05AQIEE33 pKa = 4.05YY34 pKa = 8.63WQLIRR39 pKa = 11.84KK40 pKa = 8.64EE41 pKa = 4.36SVLMYY46 pKa = 9.58YY47 pKa = 10.26GRR49 pKa = 11.84KK50 pKa = 9.56EE51 pKa = 3.82GFKK54 pKa = 11.0NFGLQPLPALTVSEE68 pKa = 4.61YY69 pKa = 10.14KK70 pKa = 10.62AKK72 pKa = 10.22EE73 pKa = 4.1AIRR76 pKa = 11.84QVLLLKK82 pKa = 10.3SLKK85 pKa = 10.62NSVFGNEE92 pKa = 3.86EE93 pKa = 3.42WTLPEE98 pKa = 3.82TSAEE102 pKa = 4.14LTYY105 pKa = 9.67TAPRR109 pKa = 11.84NTFKK113 pKa = 10.88KK114 pKa = 10.38DD115 pKa = 3.05PYY117 pKa = 10.42TVDD120 pKa = 2.01VWFDD124 pKa = 3.8HH125 pKa = 6.84NPDD128 pKa = 3.01NSFPYY133 pKa = 10.03TNWNKK138 pKa = 9.97IYY140 pKa = 10.35FQGDD144 pKa = 2.95HH145 pKa = 7.21DD146 pKa = 4.0NWHH149 pKa = 6.32RR150 pKa = 11.84AAGKK154 pKa = 10.14VDD156 pKa = 3.24INGLYY161 pKa = 10.5FEE163 pKa = 5.55DD164 pKa = 4.09ANGDD168 pKa = 2.96KK169 pKa = 10.97SYY171 pKa = 11.0FVVFASDD178 pKa = 3.11AQRR181 pKa = 11.84YY182 pKa = 5.84GTTGEE187 pKa = 4.01WTVHH191 pKa = 5.35YY192 pKa = 10.09KK193 pKa = 10.71NEE195 pKa = 4.7TISSTSLSSQRR206 pKa = 11.84SLSTISSQGSVSSSRR221 pKa = 11.84DD222 pKa = 3.31SIPAKK227 pKa = 10.05KK228 pKa = 9.88GQSTRR233 pKa = 11.84RR234 pKa = 11.84HH235 pKa = 4.56QSEE238 pKa = 4.07EE239 pKa = 4.32GIADD243 pKa = 3.7SSSSSTSSSPSAVRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84PQQGEE267 pKa = 3.94HH268 pKa = 5.46TSRR271 pKa = 11.84GGGTRR276 pKa = 11.84AKK278 pKa = 9.93RR279 pKa = 11.84QRR281 pKa = 11.84TEE283 pKa = 3.98EE284 pKa = 4.03VSTTVSPDD292 pKa = 2.85EE293 pKa = 4.15VGRR296 pKa = 11.84GNTSVPRR303 pKa = 11.84TNLTRR308 pKa = 11.84LEE310 pKa = 4.12RR311 pKa = 11.84LAKK314 pKa = 9.69EE315 pKa = 4.22ARR317 pKa = 11.84DD318 pKa = 3.72PPIVLIKK325 pKa = 10.68GRR327 pKa = 11.84SNPLKK332 pKa = 9.17CWRR335 pKa = 11.84YY336 pKa = 9.56RR337 pKa = 11.84CEE339 pKa = 4.1KK340 pKa = 10.72HH341 pKa = 6.68CDD343 pKa = 3.11LYY345 pKa = 11.68DD346 pKa = 4.97RR347 pKa = 11.84ISTVFKK353 pKa = 10.02WVNSTNDD360 pKa = 3.46CNMHH364 pKa = 6.72RR365 pKa = 11.84LLISFKK371 pKa = 10.42NQNQRR376 pKa = 11.84SMFLKK381 pKa = 10.44FVNLPKK387 pKa = 10.88GCTFSLGSLDD397 pKa = 4.62SII399 pKa = 5.04
Molecular weight: 45.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2505 |
97 |
608 |
357.9 |
40.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.511 ± 0.515 | 2.555 ± 0.861 |
6.667 ± 0.42 | 6.228 ± 0.41 |
5.03 ± 0.452 | 5.19 ± 0.609 |
1.916 ± 0.272 | 5.469 ± 0.715 |
5.629 ± 0.923 | 9.261 ± 0.975 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.756 ± 0.338 | 5.07 ± 0.405 |
5.988 ± 1.149 | 4.271 ± 0.428 |
5.788 ± 0.626 | 7.824 ± 1.019 |
5.948 ± 0.57 | 6.267 ± 0.61 |
1.158 ± 0.413 | 3.473 ± 0.552 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
