
Termite associated circular virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus termi1
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
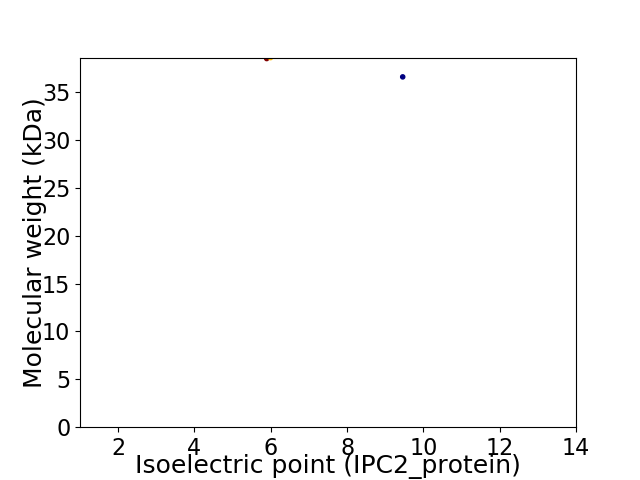
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
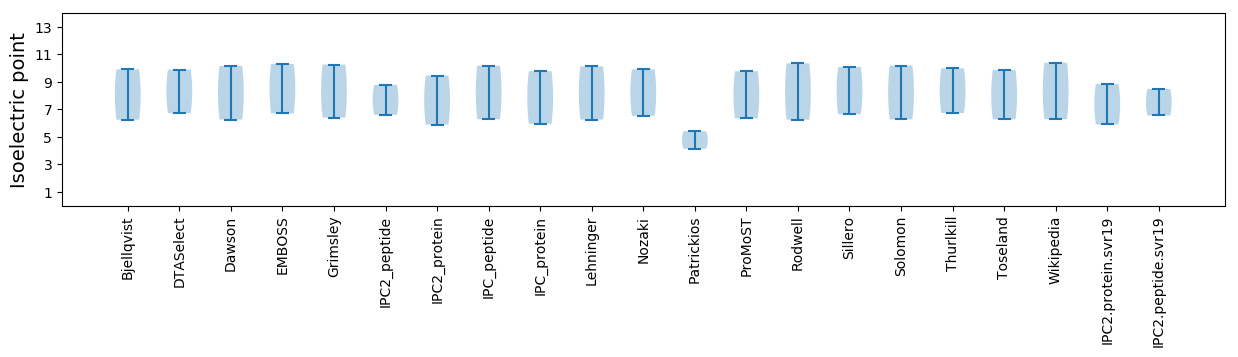
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1E314|A0A2P1E314_9VIRU Replication-associated protein OS=Termite associated circular virus 2 OX=2108550 PE=3 SV=1
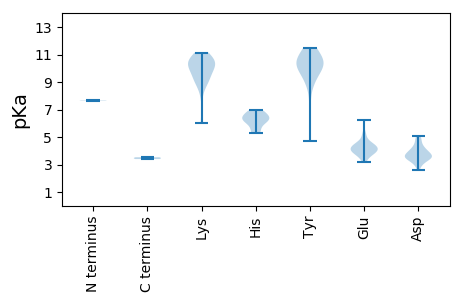
MM1 pKa = 7.68PAFVCNARR9 pKa = 11.84YY10 pKa = 8.98FLITFAQCDD19 pKa = 3.98ALDD22 pKa = 3.64PWAVSNHH29 pKa = 5.43FSTLGAEE36 pKa = 4.41CIVARR41 pKa = 11.84EE42 pKa = 4.04SHH44 pKa = 6.74ADD46 pKa = 3.43GGTHH50 pKa = 6.29LHH52 pKa = 6.48AFVDD56 pKa = 4.82FGRR59 pKa = 11.84KK60 pKa = 8.13FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84PGIFDD70 pKa = 3.79VAGCHH75 pKa = 6.08PNISPSRR82 pKa = 11.84GSPGAGYY89 pKa = 10.44DD90 pKa = 3.6YY91 pKa = 10.27ATKK94 pKa = 10.77DD95 pKa = 3.21GDD97 pKa = 3.74IVAGGLARR105 pKa = 11.84PSVDD109 pKa = 3.37AGDD112 pKa = 3.93GKK114 pKa = 10.78VDD116 pKa = 3.15KK117 pKa = 10.01WSQIISAEE125 pKa = 4.12DD126 pKa = 3.37EE127 pKa = 4.1RR128 pKa = 11.84TFWDD132 pKa = 4.0LLEE135 pKa = 4.58SLDD138 pKa = 4.22PKK140 pKa = 11.14ALATNYY146 pKa = 10.69GNLRR150 pKa = 11.84KK151 pKa = 9.87YY152 pKa = 11.13ADD154 pKa = 3.56FRR156 pKa = 11.84YY157 pKa = 9.91KK158 pKa = 10.54PEE160 pKa = 4.73PEE162 pKa = 4.56TYY164 pKa = 9.31QHH166 pKa = 6.8PAGIEE171 pKa = 3.85FGGGVLPDD179 pKa = 3.47LVAWRR184 pKa = 11.84DD185 pKa = 3.49SSLGASLGGRR195 pKa = 11.84RR196 pKa = 11.84NTSSARR202 pKa = 11.84GPPGALRR209 pKa = 11.84VYY211 pKa = 11.5AMLTSLGRR219 pKa = 11.84SRR221 pKa = 11.84SLCIYY226 pKa = 10.17GPTRR230 pKa = 11.84VGKK233 pKa = 6.01TTWARR238 pKa = 11.84SLGMHH243 pKa = 6.43VYY245 pKa = 9.93FMGMISGEE253 pKa = 3.8IAVRR257 pKa = 11.84DD258 pKa = 3.99MPTAEE263 pKa = 3.99YY264 pKa = 10.42AIFDD268 pKa = 4.19DD269 pKa = 3.8MRR271 pKa = 11.84GGIKK275 pKa = 10.07FFPAWKK281 pKa = 9.05EE282 pKa = 3.64WFGAQQVVTVKK293 pKa = 10.64KK294 pKa = 10.54LYY296 pKa = 10.2RR297 pKa = 11.84DD298 pKa = 3.48PLQINWGKK306 pKa = 8.98PCIWLSNTDD315 pKa = 3.46PRR317 pKa = 11.84TGMEE321 pKa = 4.04MEE323 pKa = 5.17DD324 pKa = 3.87IEE326 pKa = 4.4WLEE329 pKa = 4.19GNCDD333 pKa = 3.6FVFMQDD339 pKa = 4.55SIISHH344 pKa = 6.93ANTPP348 pKa = 3.53
MM1 pKa = 7.68PAFVCNARR9 pKa = 11.84YY10 pKa = 8.98FLITFAQCDD19 pKa = 3.98ALDD22 pKa = 3.64PWAVSNHH29 pKa = 5.43FSTLGAEE36 pKa = 4.41CIVARR41 pKa = 11.84EE42 pKa = 4.04SHH44 pKa = 6.74ADD46 pKa = 3.43GGTHH50 pKa = 6.29LHH52 pKa = 6.48AFVDD56 pKa = 4.82FGRR59 pKa = 11.84KK60 pKa = 8.13FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84PGIFDD70 pKa = 3.79VAGCHH75 pKa = 6.08PNISPSRR82 pKa = 11.84GSPGAGYY89 pKa = 10.44DD90 pKa = 3.6YY91 pKa = 10.27ATKK94 pKa = 10.77DD95 pKa = 3.21GDD97 pKa = 3.74IVAGGLARR105 pKa = 11.84PSVDD109 pKa = 3.37AGDD112 pKa = 3.93GKK114 pKa = 10.78VDD116 pKa = 3.15KK117 pKa = 10.01WSQIISAEE125 pKa = 4.12DD126 pKa = 3.37EE127 pKa = 4.1RR128 pKa = 11.84TFWDD132 pKa = 4.0LLEE135 pKa = 4.58SLDD138 pKa = 4.22PKK140 pKa = 11.14ALATNYY146 pKa = 10.69GNLRR150 pKa = 11.84KK151 pKa = 9.87YY152 pKa = 11.13ADD154 pKa = 3.56FRR156 pKa = 11.84YY157 pKa = 9.91KK158 pKa = 10.54PEE160 pKa = 4.73PEE162 pKa = 4.56TYY164 pKa = 9.31QHH166 pKa = 6.8PAGIEE171 pKa = 3.85FGGGVLPDD179 pKa = 3.47LVAWRR184 pKa = 11.84DD185 pKa = 3.49SSLGASLGGRR195 pKa = 11.84RR196 pKa = 11.84NTSSARR202 pKa = 11.84GPPGALRR209 pKa = 11.84VYY211 pKa = 11.5AMLTSLGRR219 pKa = 11.84SRR221 pKa = 11.84SLCIYY226 pKa = 10.17GPTRR230 pKa = 11.84VGKK233 pKa = 6.01TTWARR238 pKa = 11.84SLGMHH243 pKa = 6.43VYY245 pKa = 9.93FMGMISGEE253 pKa = 3.8IAVRR257 pKa = 11.84DD258 pKa = 3.99MPTAEE263 pKa = 3.99YY264 pKa = 10.42AIFDD268 pKa = 4.19DD269 pKa = 3.8MRR271 pKa = 11.84GGIKK275 pKa = 10.07FFPAWKK281 pKa = 9.05EE282 pKa = 3.64WFGAQQVVTVKK293 pKa = 10.64KK294 pKa = 10.54LYY296 pKa = 10.2RR297 pKa = 11.84DD298 pKa = 3.48PLQINWGKK306 pKa = 8.98PCIWLSNTDD315 pKa = 3.46PRR317 pKa = 11.84TGMEE321 pKa = 4.04MEE323 pKa = 5.17DD324 pKa = 3.87IEE326 pKa = 4.4WLEE329 pKa = 4.19GNCDD333 pKa = 3.6FVFMQDD339 pKa = 4.55SIISHH344 pKa = 6.93ANTPP348 pKa = 3.53
Molecular weight: 38.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1E314|A0A2P1E314_9VIRU Replication-associated protein OS=Termite associated circular virus 2 OX=2108550 PE=3 SV=1
MM1 pKa = 7.65AYY3 pKa = 10.05GRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84LYY9 pKa = 10.45SSRR12 pKa = 11.84VKK14 pKa = 10.08TRR16 pKa = 11.84TGISGRR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84YY26 pKa = 4.73STRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 10.21VKK33 pKa = 9.05TRR35 pKa = 11.84PLARR39 pKa = 11.84RR40 pKa = 11.84TYY42 pKa = 9.15RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84TTMSRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.23ILNISSEE60 pKa = 4.27KK61 pKa = 10.8KK62 pKa = 9.85KK63 pKa = 10.38DD64 pKa = 3.69TMLTWTNLQGSVGVEE79 pKa = 3.97TPGPAKK85 pKa = 10.26LAPPSPDD92 pKa = 3.0NLMTEE97 pKa = 4.36YY98 pKa = 10.67CVVFSPTHH106 pKa = 5.87RR107 pKa = 11.84GATFSSGSHH116 pKa = 5.28GSKK119 pKa = 10.28YY120 pKa = 10.36YY121 pKa = 10.27KK122 pKa = 9.36QLRR125 pKa = 11.84TSSLCYY131 pKa = 8.6WRR133 pKa = 11.84GIRR136 pKa = 11.84EE137 pKa = 4.28VVTLKK142 pKa = 10.17TDD144 pKa = 4.01GPGAWEE150 pKa = 3.64WRR152 pKa = 11.84RR153 pKa = 11.84ICFTMKK159 pKa = 10.9GEE161 pKa = 4.27DD162 pKa = 4.24LYY164 pKa = 11.31TGDD167 pKa = 4.86RR168 pKa = 11.84YY169 pKa = 11.32SFDD172 pKa = 4.4LGTSPVARR180 pKa = 11.84ITSDD184 pKa = 2.62GMKK187 pKa = 10.03RR188 pKa = 11.84AFFSSEE194 pKa = 3.35NALRR198 pKa = 11.84TRR200 pKa = 11.84DD201 pKa = 3.85AIFDD205 pKa = 3.77GNRR208 pKa = 11.84NVDD211 pKa = 3.1WADD214 pKa = 3.48VITAKK219 pKa = 10.46VDD221 pKa = 3.5TEE223 pKa = 5.21LITPLYY229 pKa = 9.36DD230 pKa = 2.9RR231 pKa = 11.84RR232 pKa = 11.84MLIRR236 pKa = 11.84SGNEE240 pKa = 3.17AGTVRR245 pKa = 11.84IKK247 pKa = 10.84KK248 pKa = 9.31CWHH251 pKa = 6.27AMNKK255 pKa = 8.78NLQYY259 pKa = 11.45DD260 pKa = 3.51DD261 pKa = 5.08DD262 pKa = 4.88EE263 pKa = 6.21NGEE266 pKa = 4.45SMLEE270 pKa = 3.86RR271 pKa = 11.84YY272 pKa = 9.72YY273 pKa = 10.45STEE276 pKa = 3.87GKK278 pKa = 9.54PGMGDD283 pKa = 3.45YY284 pKa = 11.08YY285 pKa = 10.43IVDD288 pKa = 4.39LFQAVVPQQNVTPTNLYY305 pKa = 10.74VNIEE309 pKa = 3.92ATAYY313 pKa = 7.14WHH315 pKa = 6.3EE316 pKa = 4.26RR317 pKa = 3.4
MM1 pKa = 7.65AYY3 pKa = 10.05GRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84LYY9 pKa = 10.45SSRR12 pKa = 11.84VKK14 pKa = 10.08TRR16 pKa = 11.84TGISGRR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84YY26 pKa = 4.73STRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 10.21VKK33 pKa = 9.05TRR35 pKa = 11.84PLARR39 pKa = 11.84RR40 pKa = 11.84TYY42 pKa = 9.15RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84TTMSRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.23ILNISSEE60 pKa = 4.27KK61 pKa = 10.8KK62 pKa = 9.85KK63 pKa = 10.38DD64 pKa = 3.69TMLTWTNLQGSVGVEE79 pKa = 3.97TPGPAKK85 pKa = 10.26LAPPSPDD92 pKa = 3.0NLMTEE97 pKa = 4.36YY98 pKa = 10.67CVVFSPTHH106 pKa = 5.87RR107 pKa = 11.84GATFSSGSHH116 pKa = 5.28GSKK119 pKa = 10.28YY120 pKa = 10.36YY121 pKa = 10.27KK122 pKa = 9.36QLRR125 pKa = 11.84TSSLCYY131 pKa = 8.6WRR133 pKa = 11.84GIRR136 pKa = 11.84EE137 pKa = 4.28VVTLKK142 pKa = 10.17TDD144 pKa = 4.01GPGAWEE150 pKa = 3.64WRR152 pKa = 11.84RR153 pKa = 11.84ICFTMKK159 pKa = 10.9GEE161 pKa = 4.27DD162 pKa = 4.24LYY164 pKa = 11.31TGDD167 pKa = 4.86RR168 pKa = 11.84YY169 pKa = 11.32SFDD172 pKa = 4.4LGTSPVARR180 pKa = 11.84ITSDD184 pKa = 2.62GMKK187 pKa = 10.03RR188 pKa = 11.84AFFSSEE194 pKa = 3.35NALRR198 pKa = 11.84TRR200 pKa = 11.84DD201 pKa = 3.85AIFDD205 pKa = 3.77GNRR208 pKa = 11.84NVDD211 pKa = 3.1WADD214 pKa = 3.48VITAKK219 pKa = 10.46VDD221 pKa = 3.5TEE223 pKa = 5.21LITPLYY229 pKa = 9.36DD230 pKa = 2.9RR231 pKa = 11.84RR232 pKa = 11.84MLIRR236 pKa = 11.84SGNEE240 pKa = 3.17AGTVRR245 pKa = 11.84IKK247 pKa = 10.84KK248 pKa = 9.31CWHH251 pKa = 6.27AMNKK255 pKa = 8.78NLQYY259 pKa = 11.45DD260 pKa = 3.51DD261 pKa = 5.08DD262 pKa = 4.88EE263 pKa = 6.21NGEE266 pKa = 4.45SMLEE270 pKa = 3.86RR271 pKa = 11.84YY272 pKa = 9.72YY273 pKa = 10.45STEE276 pKa = 3.87GKK278 pKa = 9.54PGMGDD283 pKa = 3.45YY284 pKa = 11.08YY285 pKa = 10.43IVDD288 pKa = 4.39LFQAVVPQQNVTPTNLYY305 pKa = 10.74VNIEE309 pKa = 3.92ATAYY313 pKa = 7.14WHH315 pKa = 6.3EE316 pKa = 4.26RR317 pKa = 3.4
Molecular weight: 36.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
665 |
317 |
348 |
332.5 |
37.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.368 ± 1.301 | 1.654 ± 0.255 |
6.466 ± 0.511 | 4.511 ± 0.143 |
3.91 ± 0.899 | 9.023 ± 0.942 |
1.805 ± 0.352 | 4.662 ± 0.364 |
4.511 ± 0.552 | 6.466 ± 0.102 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.008 ± 0.095 | 3.459 ± 0.417 |
5.113 ± 0.657 | 1.955 ± 0.04 |
9.173 ± 1.621 | 7.218 ± 0.229 |
7.068 ± 1.555 | 5.414 ± 0.172 |
2.556 ± 0.226 | 4.662 ± 0.864 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
