
Gallid herpesvirus 2 (strain Chicken/Md5/ATCC VR-987) (GaHV-2) (Marek s disease herpesvirus type 1)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Mardivirus; Gallid alphaherpesvirus 2
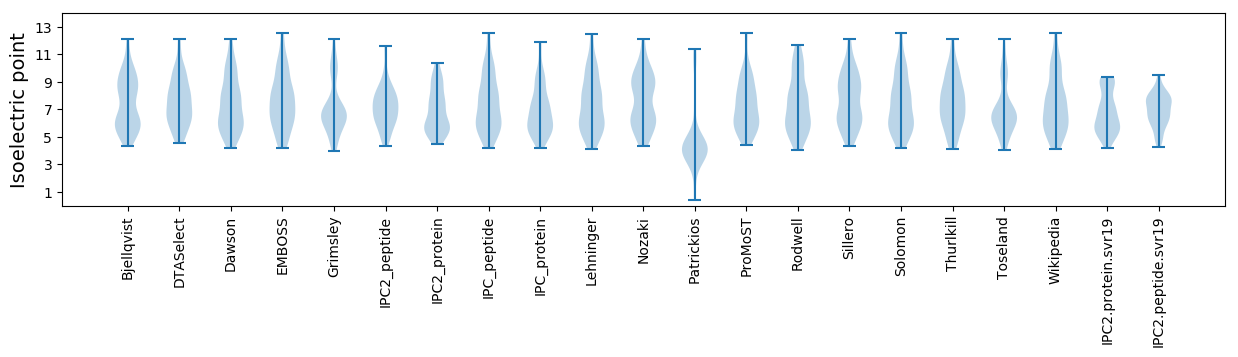
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
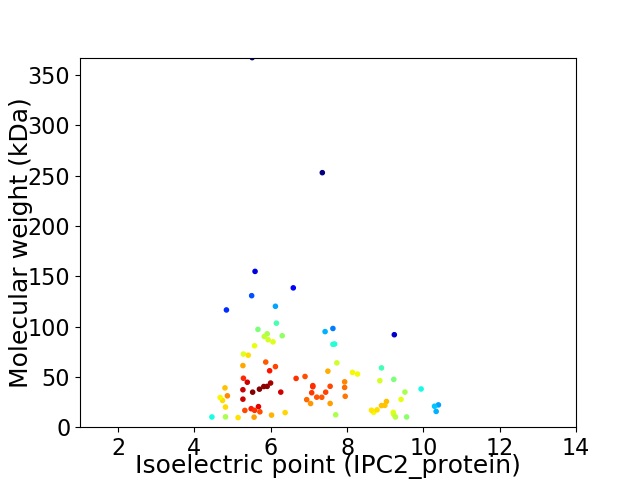
Virtual 2D-PAGE plot for 91 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q77MR5|VP16_GAHVM Tegument protein VP16 homolog OS=Gallid herpesvirus 2 (strain Chicken/Md5/ATCC VR-987) OX=10389 GN=MDV061 PE=3 SV=1
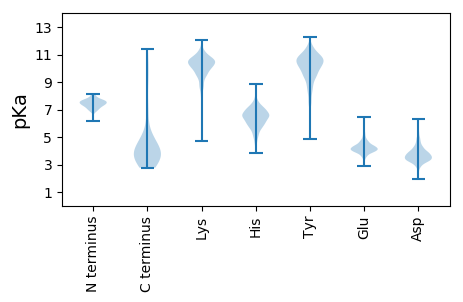
MM1 pKa = 7.58GLMDD5 pKa = 3.88IHH7 pKa = 6.54NAVCSLVIGVAILIATSQATFVDD30 pKa = 3.87WGSSITSMGDD40 pKa = 2.94FWEE43 pKa = 4.92STCSAVGVSIAFSSGFSVLFYY64 pKa = 9.71MGLVAVISALLAGSYY79 pKa = 8.93HH80 pKa = 5.92ACFRR84 pKa = 11.84LFTADD89 pKa = 3.19MFKK92 pKa = 11.09EE93 pKa = 4.32EE94 pKa = 4.02WW95 pKa = 3.25
MM1 pKa = 7.58GLMDD5 pKa = 3.88IHH7 pKa = 6.54NAVCSLVIGVAILIATSQATFVDD30 pKa = 3.87WGSSITSMGDD40 pKa = 2.94FWEE43 pKa = 4.92STCSAVGVSIAFSSGFSVLFYY64 pKa = 9.71MGLVAVISALLAGSYY79 pKa = 8.93HH80 pKa = 5.92ACFRR84 pKa = 11.84LFTADD89 pKa = 3.19MFKK92 pKa = 11.09EE93 pKa = 4.32EE94 pKa = 4.02WW95 pKa = 3.25
Molecular weight: 10.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9E6P5|KITH_GAHVM Thymidine kinase OS=Gallid herpesvirus 2 (strain Chicken/Md5/ATCC VR-987) OX=10389 GN=TK PE=3 SV=1
MM1 pKa = 7.53SSEE4 pKa = 4.29MPLPTTIVAPTCSMGEE20 pKa = 4.01KK21 pKa = 10.18NVLKK25 pKa = 10.33RR26 pKa = 11.84YY27 pKa = 8.91RR28 pKa = 11.84RR29 pKa = 11.84QVSSTRR35 pKa = 11.84NFKK38 pKa = 10.58RR39 pKa = 11.84HH40 pKa = 4.74VNTNVLRR47 pKa = 11.84KK48 pKa = 9.12RR49 pKa = 11.84RR50 pKa = 11.84LAAGVRR56 pKa = 11.84CHH58 pKa = 6.91DD59 pKa = 3.41RR60 pKa = 11.84FYY62 pKa = 11.5KK63 pKa = 10.3RR64 pKa = 11.84LYY66 pKa = 10.22AEE68 pKa = 4.41AMCLGSQVYY77 pKa = 9.4DD78 pKa = 3.2WPGRR82 pKa = 11.84SFAKK86 pKa = 10.31IFGKK90 pKa = 10.93VMVLDD95 pKa = 3.81VFKK98 pKa = 11.06QLTDD102 pKa = 3.64FRR104 pKa = 11.84LVFEE108 pKa = 4.72VNLEE112 pKa = 3.88RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84PDD117 pKa = 4.09CICMFKK123 pKa = 10.72LPRR126 pKa = 11.84DD127 pKa = 3.6LWEE130 pKa = 4.18VGCDD134 pKa = 3.72GVCVILEE141 pKa = 4.17LKK143 pKa = 8.3TCKK146 pKa = 10.03FSRR149 pKa = 11.84NLKK152 pKa = 8.43TRR154 pKa = 11.84SKK156 pKa = 10.7HH157 pKa = 4.12EE158 pKa = 3.8QRR160 pKa = 11.84LTGIKK165 pKa = 9.8QLLDD169 pKa = 3.61SKK171 pKa = 11.32SLIGQIAPQGSDD183 pKa = 4.05CIVICPMLVFVARR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 10.36LSVLHH203 pKa = 6.09VMCLKK208 pKa = 10.19KK209 pKa = 10.54RR210 pKa = 11.84HH211 pKa = 5.93IVTDD215 pKa = 4.1FHH217 pKa = 6.89RR218 pKa = 11.84LCSILATASDD228 pKa = 3.98YY229 pKa = 10.86KK230 pKa = 11.33ARR232 pKa = 11.84ITDD235 pKa = 3.07THH237 pKa = 6.84KK238 pKa = 10.25ILKK241 pKa = 9.44RR242 pKa = 11.84HH243 pKa = 5.08NTNTIHH249 pKa = 6.07IQRR252 pKa = 11.84RR253 pKa = 11.84SGKK256 pKa = 10.14ASANVTDD263 pKa = 4.46LRR265 pKa = 11.84NHH267 pKa = 6.44IACNKK272 pKa = 7.38TATLSFSNNQEE283 pKa = 4.14RR284 pKa = 11.84TGGTAMRR291 pKa = 11.84NMANIIARR299 pKa = 11.84LVQRR303 pKa = 11.84PP304 pKa = 3.28
MM1 pKa = 7.53SSEE4 pKa = 4.29MPLPTTIVAPTCSMGEE20 pKa = 4.01KK21 pKa = 10.18NVLKK25 pKa = 10.33RR26 pKa = 11.84YY27 pKa = 8.91RR28 pKa = 11.84RR29 pKa = 11.84QVSSTRR35 pKa = 11.84NFKK38 pKa = 10.58RR39 pKa = 11.84HH40 pKa = 4.74VNTNVLRR47 pKa = 11.84KK48 pKa = 9.12RR49 pKa = 11.84RR50 pKa = 11.84LAAGVRR56 pKa = 11.84CHH58 pKa = 6.91DD59 pKa = 3.41RR60 pKa = 11.84FYY62 pKa = 11.5KK63 pKa = 10.3RR64 pKa = 11.84LYY66 pKa = 10.22AEE68 pKa = 4.41AMCLGSQVYY77 pKa = 9.4DD78 pKa = 3.2WPGRR82 pKa = 11.84SFAKK86 pKa = 10.31IFGKK90 pKa = 10.93VMVLDD95 pKa = 3.81VFKK98 pKa = 11.06QLTDD102 pKa = 3.64FRR104 pKa = 11.84LVFEE108 pKa = 4.72VNLEE112 pKa = 3.88RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84PDD117 pKa = 4.09CICMFKK123 pKa = 10.72LPRR126 pKa = 11.84DD127 pKa = 3.6LWEE130 pKa = 4.18VGCDD134 pKa = 3.72GVCVILEE141 pKa = 4.17LKK143 pKa = 8.3TCKK146 pKa = 10.03FSRR149 pKa = 11.84NLKK152 pKa = 8.43TRR154 pKa = 11.84SKK156 pKa = 10.7HH157 pKa = 4.12EE158 pKa = 3.8QRR160 pKa = 11.84LTGIKK165 pKa = 9.8QLLDD169 pKa = 3.61SKK171 pKa = 11.32SLIGQIAPQGSDD183 pKa = 4.05CIVICPMLVFVARR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 10.36LSVLHH203 pKa = 6.09VMCLKK208 pKa = 10.19KK209 pKa = 10.54RR210 pKa = 11.84HH211 pKa = 5.93IVTDD215 pKa = 4.1FHH217 pKa = 6.89RR218 pKa = 11.84LCSILATASDD228 pKa = 3.98YY229 pKa = 10.86KK230 pKa = 11.33ARR232 pKa = 11.84ITDD235 pKa = 3.07THH237 pKa = 6.84KK238 pKa = 10.25ILKK241 pKa = 9.44RR242 pKa = 11.84HH243 pKa = 5.08NTNTIHH249 pKa = 6.07IQRR252 pKa = 11.84RR253 pKa = 11.84SGKK256 pKa = 10.14ASANVTDD263 pKa = 4.46LRR265 pKa = 11.84NHH267 pKa = 6.44IACNKK272 pKa = 7.38TATLSFSNNQEE283 pKa = 4.14RR284 pKa = 11.84TGGTAMRR291 pKa = 11.84NMANIIARR299 pKa = 11.84LVQRR303 pKa = 11.84PP304 pKa = 3.28
Molecular weight: 34.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
42836 |
84 |
3342 |
470.7 |
52.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.169 ± 0.187 | 2.386 ± 0.136 |
5.507 ± 0.167 | 5.355 ± 0.153 |
3.803 ± 0.176 | 5.619 ± 0.174 |
2.493 ± 0.08 | 6.546 ± 0.245 |
4.249 ± 0.196 | 9.205 ± 0.187 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.126 | 4.337 ± 0.156 |
5.57 ± 0.506 | 2.685 ± 0.1 |
6.665 ± 0.253 | 8.635 ± 0.285 |
6.695 ± 0.143 | 5.843 ± 0.14 |
1.137 ± 0.073 | 3.497 ± 0.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
