
Lake Sarah-associated circular virus-2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
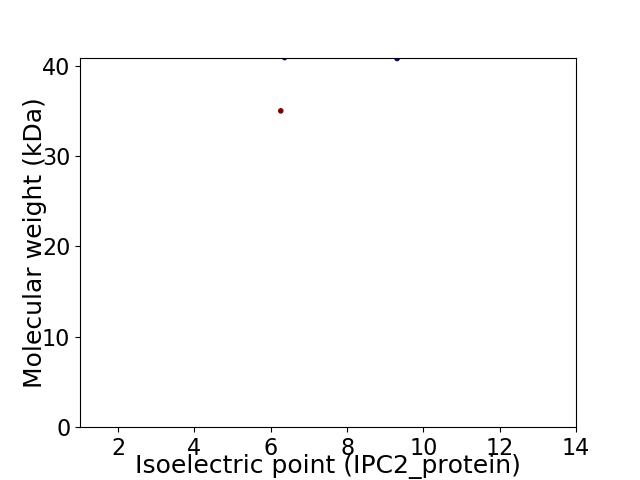
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
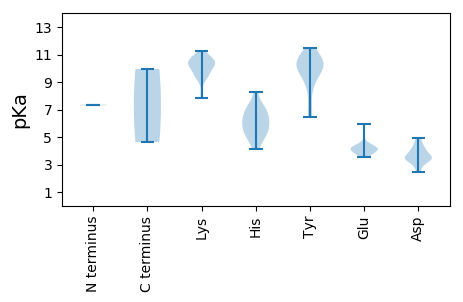
Protein with the lowest isoelectric point:
>tr|A0A140AQJ4|A0A140AQJ4_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-2 OX=1685746 PE=3 SV=1
MM1 pKa = 7.35QINAKK6 pKa = 10.16SFFLTYY12 pKa = 9.29PRR14 pKa = 11.84CAITAEE20 pKa = 4.34TFQLWLEE27 pKa = 4.37STFTPTYY34 pKa = 10.47LVIGSEE40 pKa = 4.15LHH42 pKa = 6.27QDD44 pKa = 3.83GTPHH48 pKa = 5.28VHH50 pKa = 6.07VCFTLDD56 pKa = 2.94TALRR60 pKa = 11.84TRR62 pKa = 11.84DD63 pKa = 3.26EE64 pKa = 4.34RR65 pKa = 11.84VFDD68 pKa = 3.97FHH70 pKa = 6.8GHH72 pKa = 6.01HH73 pKa = 7.05PNIVRR78 pKa = 11.84PRR80 pKa = 11.84AIKK83 pKa = 10.35KK84 pKa = 10.36CIDD87 pKa = 3.48YY88 pKa = 10.84VKK90 pKa = 10.73KK91 pKa = 10.57DD92 pKa = 3.44GNFIEE97 pKa = 5.19SGEE100 pKa = 4.04PPAMKK105 pKa = 10.22RR106 pKa = 11.84KK107 pKa = 8.94WSEE110 pKa = 3.68LQTATTSEE118 pKa = 4.38EE119 pKa = 4.07FFKK122 pKa = 10.57TALEE126 pKa = 3.93ISPRR130 pKa = 11.84DD131 pKa = 3.38YY132 pKa = 11.48YY133 pKa = 11.48LNQEE137 pKa = 4.12RR138 pKa = 11.84LEE140 pKa = 4.07YY141 pKa = 9.45MARR144 pKa = 11.84KK145 pKa = 9.18LFKK148 pKa = 10.12PVVPTYY154 pKa = 10.32VPQWVDD160 pKa = 3.75FNIPQEE166 pKa = 4.05LSDD169 pKa = 4.65WIDD172 pKa = 3.29QRR174 pKa = 11.84HH175 pKa = 4.61EE176 pKa = 3.75VCYY179 pKa = 10.33RR180 pKa = 11.84PQSLILYY187 pKa = 7.07GASRR191 pKa = 11.84TGKK194 pKa = 7.82TEE196 pKa = 3.71WARR199 pKa = 11.84SLGPHH204 pKa = 6.33VYY206 pKa = 10.2FNGYY210 pKa = 10.11FMLDD214 pKa = 4.12LIRR217 pKa = 11.84DD218 pKa = 3.89DD219 pKa = 3.52VEE221 pKa = 3.94YY222 pKa = 10.83AIFDD226 pKa = 4.23DD227 pKa = 4.94FEE229 pKa = 5.96DD230 pKa = 3.34WTTFKK235 pKa = 10.67QYY237 pKa = 10.67KK238 pKa = 8.18QWLGAQKK245 pKa = 10.53QFVCTDD251 pKa = 3.51KK252 pKa = 11.09YY253 pKa = 10.63RR254 pKa = 11.84KK255 pKa = 9.78KK256 pKa = 10.94VDD258 pKa = 4.08LLWGKK263 pKa = 9.36PCIILSNEE271 pKa = 3.55YY272 pKa = 9.83PAFRR276 pKa = 11.84DD277 pKa = 3.54QDD279 pKa = 4.08WILLNCITVKK289 pKa = 10.58LNKK292 pKa = 9.39PLYY295 pKa = 9.95
MM1 pKa = 7.35QINAKK6 pKa = 10.16SFFLTYY12 pKa = 9.29PRR14 pKa = 11.84CAITAEE20 pKa = 4.34TFQLWLEE27 pKa = 4.37STFTPTYY34 pKa = 10.47LVIGSEE40 pKa = 4.15LHH42 pKa = 6.27QDD44 pKa = 3.83GTPHH48 pKa = 5.28VHH50 pKa = 6.07VCFTLDD56 pKa = 2.94TALRR60 pKa = 11.84TRR62 pKa = 11.84DD63 pKa = 3.26EE64 pKa = 4.34RR65 pKa = 11.84VFDD68 pKa = 3.97FHH70 pKa = 6.8GHH72 pKa = 6.01HH73 pKa = 7.05PNIVRR78 pKa = 11.84PRR80 pKa = 11.84AIKK83 pKa = 10.35KK84 pKa = 10.36CIDD87 pKa = 3.48YY88 pKa = 10.84VKK90 pKa = 10.73KK91 pKa = 10.57DD92 pKa = 3.44GNFIEE97 pKa = 5.19SGEE100 pKa = 4.04PPAMKK105 pKa = 10.22RR106 pKa = 11.84KK107 pKa = 8.94WSEE110 pKa = 3.68LQTATTSEE118 pKa = 4.38EE119 pKa = 4.07FFKK122 pKa = 10.57TALEE126 pKa = 3.93ISPRR130 pKa = 11.84DD131 pKa = 3.38YY132 pKa = 11.48YY133 pKa = 11.48LNQEE137 pKa = 4.12RR138 pKa = 11.84LEE140 pKa = 4.07YY141 pKa = 9.45MARR144 pKa = 11.84KK145 pKa = 9.18LFKK148 pKa = 10.12PVVPTYY154 pKa = 10.32VPQWVDD160 pKa = 3.75FNIPQEE166 pKa = 4.05LSDD169 pKa = 4.65WIDD172 pKa = 3.29QRR174 pKa = 11.84HH175 pKa = 4.61EE176 pKa = 3.75VCYY179 pKa = 10.33RR180 pKa = 11.84PQSLILYY187 pKa = 7.07GASRR191 pKa = 11.84TGKK194 pKa = 7.82TEE196 pKa = 3.71WARR199 pKa = 11.84SLGPHH204 pKa = 6.33VYY206 pKa = 10.2FNGYY210 pKa = 10.11FMLDD214 pKa = 4.12LIRR217 pKa = 11.84DD218 pKa = 3.89DD219 pKa = 3.52VEE221 pKa = 3.94YY222 pKa = 10.83AIFDD226 pKa = 4.23DD227 pKa = 4.94FEE229 pKa = 5.96DD230 pKa = 3.34WTTFKK235 pKa = 10.67QYY237 pKa = 10.67KK238 pKa = 8.18QWLGAQKK245 pKa = 10.53QFVCTDD251 pKa = 3.51KK252 pKa = 11.09YY253 pKa = 10.63RR254 pKa = 11.84KK255 pKa = 9.78KK256 pKa = 10.94VDD258 pKa = 4.08LLWGKK263 pKa = 9.36PCIILSNEE271 pKa = 3.55YY272 pKa = 9.83PAFRR276 pKa = 11.84DD277 pKa = 3.54QDD279 pKa = 4.08WILLNCITVKK289 pKa = 10.58LNKK292 pKa = 9.39PLYY295 pKa = 9.95
Molecular weight: 35.0 kDa
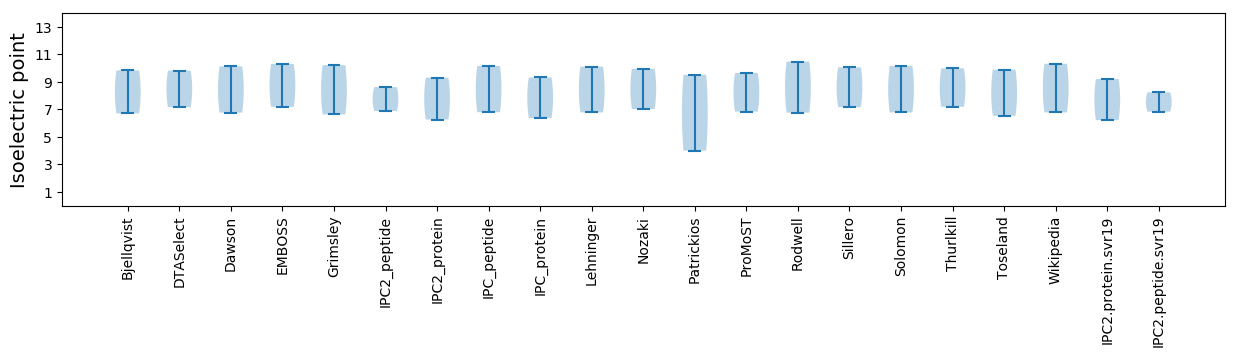
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQJ4|A0A140AQJ4_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-2 OX=1685746 PE=3 SV=1
MM1 pKa = 7.3PRR3 pKa = 11.84SVSRR7 pKa = 11.84GRR9 pKa = 11.84SVSRR13 pKa = 11.84GSGAYY18 pKa = 6.43PTPSGKK24 pKa = 10.13RR25 pKa = 11.84GGPSGKK31 pKa = 9.35RR32 pKa = 11.84GRR34 pKa = 11.84STTRR38 pKa = 11.84SSKK41 pKa = 10.66GKK43 pKa = 9.42SKK45 pKa = 9.53RR46 pKa = 11.84TKK48 pKa = 9.46RR49 pKa = 11.84SHH51 pKa = 5.42SRR53 pKa = 11.84SRR55 pKa = 11.84SHH57 pKa = 7.15RR58 pKa = 11.84GKK60 pKa = 9.96HH61 pKa = 5.04VYY63 pKa = 9.09NPSMIAGGRR72 pKa = 11.84SQAVIKK78 pKa = 10.5LGKK81 pKa = 9.77DD82 pKa = 2.74KK83 pKa = 10.95KK84 pKa = 10.64LPKK87 pKa = 10.1FMKK90 pKa = 10.21FVSQPITHH98 pKa = 7.34IDD100 pKa = 3.12TGAVIVKK107 pKa = 10.04DD108 pKa = 3.99VAGKK112 pKa = 9.73AAYY115 pKa = 9.65SYY117 pKa = 10.88HH118 pKa = 6.54EE119 pKa = 4.43LMHH122 pKa = 5.52YY123 pKa = 10.18TDD125 pKa = 4.73MNRR128 pKa = 11.84VWLAAARR135 pKa = 11.84ANRR138 pKa = 11.84ATVVVGGADD147 pKa = 3.43VPKK150 pKa = 9.73WDD152 pKa = 4.66EE153 pKa = 4.11KK154 pKa = 10.27NHH156 pKa = 7.08SIFLQDD162 pKa = 2.47GHH164 pKa = 8.28INIRR168 pKa = 11.84MVNGAVVTACVEE180 pKa = 4.04LFVLSTRR187 pKa = 11.84RR188 pKa = 11.84DD189 pKa = 3.73CKK191 pKa = 10.21IDD193 pKa = 3.4PVNAIGQDD201 pKa = 3.47VADD204 pKa = 4.36LNRR207 pKa = 11.84YY208 pKa = 8.4NISGTALDD216 pKa = 4.33GQNVAYY222 pKa = 10.0YY223 pKa = 9.71PSDD226 pKa = 3.38SPTFMAHH233 pKa = 4.99FHH235 pKa = 5.76VVQKK239 pKa = 10.99YY240 pKa = 6.88QMCLAPGEE248 pKa = 4.21THH250 pKa = 5.48MQNIYY255 pKa = 10.62VSWEE259 pKa = 3.72KK260 pKa = 10.3MLPTSLASSSTLTTTDD276 pKa = 3.46TFFKK280 pKa = 10.73GYY282 pKa = 8.41TCFLMYY288 pKa = 10.27RR289 pKa = 11.84IVGFPEE295 pKa = 4.28GQVGSSTACASGITDD310 pKa = 3.33CHH312 pKa = 4.12VTIDD316 pKa = 3.02KK317 pKa = 10.5RR318 pKa = 11.84FRR320 pKa = 11.84YY321 pKa = 9.52RR322 pKa = 11.84LLANDD327 pKa = 3.58SKK329 pKa = 11.23HH330 pKa = 6.33YY331 pKa = 10.2EE332 pKa = 4.18VYY334 pKa = 10.93NSVSVPGLPSFVDD347 pKa = 3.72EE348 pKa = 4.23LTGAVMRR355 pKa = 11.84TDD357 pKa = 2.66IFGNVTAAAIQTLIAA372 pKa = 4.62
MM1 pKa = 7.3PRR3 pKa = 11.84SVSRR7 pKa = 11.84GRR9 pKa = 11.84SVSRR13 pKa = 11.84GSGAYY18 pKa = 6.43PTPSGKK24 pKa = 10.13RR25 pKa = 11.84GGPSGKK31 pKa = 9.35RR32 pKa = 11.84GRR34 pKa = 11.84STTRR38 pKa = 11.84SSKK41 pKa = 10.66GKK43 pKa = 9.42SKK45 pKa = 9.53RR46 pKa = 11.84TKK48 pKa = 9.46RR49 pKa = 11.84SHH51 pKa = 5.42SRR53 pKa = 11.84SRR55 pKa = 11.84SHH57 pKa = 7.15RR58 pKa = 11.84GKK60 pKa = 9.96HH61 pKa = 5.04VYY63 pKa = 9.09NPSMIAGGRR72 pKa = 11.84SQAVIKK78 pKa = 10.5LGKK81 pKa = 9.77DD82 pKa = 2.74KK83 pKa = 10.95KK84 pKa = 10.64LPKK87 pKa = 10.1FMKK90 pKa = 10.21FVSQPITHH98 pKa = 7.34IDD100 pKa = 3.12TGAVIVKK107 pKa = 10.04DD108 pKa = 3.99VAGKK112 pKa = 9.73AAYY115 pKa = 9.65SYY117 pKa = 10.88HH118 pKa = 6.54EE119 pKa = 4.43LMHH122 pKa = 5.52YY123 pKa = 10.18TDD125 pKa = 4.73MNRR128 pKa = 11.84VWLAAARR135 pKa = 11.84ANRR138 pKa = 11.84ATVVVGGADD147 pKa = 3.43VPKK150 pKa = 9.73WDD152 pKa = 4.66EE153 pKa = 4.11KK154 pKa = 10.27NHH156 pKa = 7.08SIFLQDD162 pKa = 2.47GHH164 pKa = 8.28INIRR168 pKa = 11.84MVNGAVVTACVEE180 pKa = 4.04LFVLSTRR187 pKa = 11.84RR188 pKa = 11.84DD189 pKa = 3.73CKK191 pKa = 10.21IDD193 pKa = 3.4PVNAIGQDD201 pKa = 3.47VADD204 pKa = 4.36LNRR207 pKa = 11.84YY208 pKa = 8.4NISGTALDD216 pKa = 4.33GQNVAYY222 pKa = 10.0YY223 pKa = 9.71PSDD226 pKa = 3.38SPTFMAHH233 pKa = 4.99FHH235 pKa = 5.76VVQKK239 pKa = 10.99YY240 pKa = 6.88QMCLAPGEE248 pKa = 4.21THH250 pKa = 5.48MQNIYY255 pKa = 10.62VSWEE259 pKa = 3.72KK260 pKa = 10.3MLPTSLASSSTLTTTDD276 pKa = 3.46TFFKK280 pKa = 10.73GYY282 pKa = 8.41TCFLMYY288 pKa = 10.27RR289 pKa = 11.84IVGFPEE295 pKa = 4.28GQVGSSTACASGITDD310 pKa = 3.33CHH312 pKa = 4.12VTIDD316 pKa = 3.02KK317 pKa = 10.5RR318 pKa = 11.84FRR320 pKa = 11.84YY321 pKa = 9.52RR322 pKa = 11.84LLANDD327 pKa = 3.58SKK329 pKa = 11.23HH330 pKa = 6.33YY331 pKa = 10.2EE332 pKa = 4.18VYY334 pKa = 10.93NSVSVPGLPSFVDD347 pKa = 3.72EE348 pKa = 4.23LTGAVMRR355 pKa = 11.84TDD357 pKa = 2.66IFGNVTAAAIQTLIAA372 pKa = 4.62
Molecular weight: 40.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
667 |
295 |
372 |
333.5 |
37.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.597 ± 1.179 | 1.949 ± 0.27 |
5.847 ± 0.594 | 3.898 ± 1.404 |
4.798 ± 1.047 | 6.297 ± 1.636 |
3.148 ± 0.278 | 5.247 ± 0.328 |
6.447 ± 0.212 | 6.747 ± 1.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.399 ± 0.664 | 3.448 ± 0.253 |
4.948 ± 0.519 | 3.598 ± 0.731 |
6.447 ± 0.436 | 7.196 ± 1.993 |
7.196 ± 0.05 | 7.346 ± 1.225 |
1.799 ± 0.797 | 4.648 ± 0.494 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
