
Hubei mosquito virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
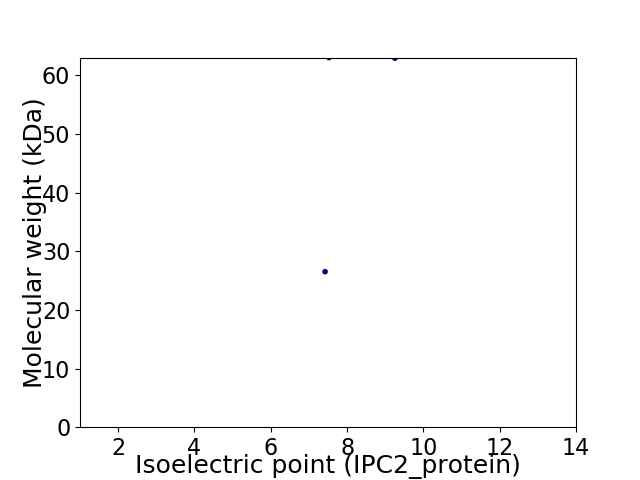
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHU0|A0A1L3KHU0_9VIRU Uncharacterized protein OS=Hubei mosquito virus 3 OX=1922927 PE=4 SV=1
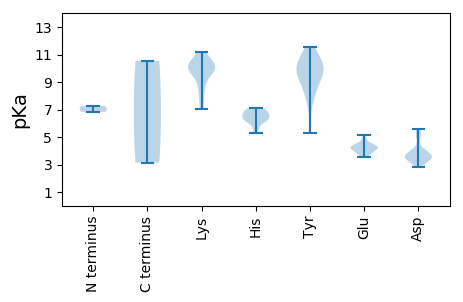
MM1 pKa = 6.83QRR3 pKa = 11.84DD4 pKa = 3.53MSGRR8 pKa = 11.84RR9 pKa = 11.84TPEE12 pKa = 4.22GSCSEE17 pKa = 4.05PSRR20 pKa = 11.84ARR22 pKa = 11.84FSVPPCEE29 pKa = 4.81PPDD32 pKa = 3.79GFCPVVQGSTLSKK45 pKa = 9.75TRR47 pKa = 11.84RR48 pKa = 11.84QTLRR52 pKa = 11.84GWITCRR58 pKa = 11.84PCGGIDD64 pKa = 3.0TSMILVLHH72 pKa = 5.56VQHH75 pKa = 6.69SKK77 pKa = 11.08LVFCSLSCEE86 pKa = 3.91MRR88 pKa = 11.84LCLSQGTATWNVYY101 pKa = 7.8VVYY104 pKa = 10.52LPAQASDD111 pKa = 3.63FSRR114 pKa = 11.84TIQGAEE120 pKa = 3.59FPMRR124 pKa = 11.84IEE126 pKa = 4.16AVPFIDD132 pKa = 3.55VTEE135 pKa = 4.36LLGSPIVVLPRR146 pKa = 11.84HH147 pKa = 6.4RR148 pKa = 11.84GQDD151 pKa = 3.37SASVEE156 pKa = 4.23MFSPNHH162 pKa = 6.2LGAEE166 pKa = 4.43CAEE169 pKa = 4.33LPGVEE174 pKa = 4.63CFPGGLHH181 pKa = 6.2EE182 pKa = 4.93AQFCGIIRR190 pKa = 11.84PPLLLRR196 pKa = 11.84VGGVCQSIVTAHH208 pKa = 6.52NEE210 pKa = 3.77FLHH213 pKa = 7.04AGLSDD218 pKa = 4.31DD219 pKa = 5.26RR220 pKa = 11.84GIPQAKK226 pKa = 8.98KK227 pKa = 9.96RR228 pKa = 11.84KK229 pKa = 9.03RR230 pKa = 11.84DD231 pKa = 3.53QGPHH235 pKa = 6.57KK236 pKa = 10.43GPPALPKK243 pKa = 10.53
MM1 pKa = 6.83QRR3 pKa = 11.84DD4 pKa = 3.53MSGRR8 pKa = 11.84RR9 pKa = 11.84TPEE12 pKa = 4.22GSCSEE17 pKa = 4.05PSRR20 pKa = 11.84ARR22 pKa = 11.84FSVPPCEE29 pKa = 4.81PPDD32 pKa = 3.79GFCPVVQGSTLSKK45 pKa = 9.75TRR47 pKa = 11.84RR48 pKa = 11.84QTLRR52 pKa = 11.84GWITCRR58 pKa = 11.84PCGGIDD64 pKa = 3.0TSMILVLHH72 pKa = 5.56VQHH75 pKa = 6.69SKK77 pKa = 11.08LVFCSLSCEE86 pKa = 3.91MRR88 pKa = 11.84LCLSQGTATWNVYY101 pKa = 7.8VVYY104 pKa = 10.52LPAQASDD111 pKa = 3.63FSRR114 pKa = 11.84TIQGAEE120 pKa = 3.59FPMRR124 pKa = 11.84IEE126 pKa = 4.16AVPFIDD132 pKa = 3.55VTEE135 pKa = 4.36LLGSPIVVLPRR146 pKa = 11.84HH147 pKa = 6.4RR148 pKa = 11.84GQDD151 pKa = 3.37SASVEE156 pKa = 4.23MFSPNHH162 pKa = 6.2LGAEE166 pKa = 4.43CAEE169 pKa = 4.33LPGVEE174 pKa = 4.63CFPGGLHH181 pKa = 6.2EE182 pKa = 4.93AQFCGIIRR190 pKa = 11.84PPLLLRR196 pKa = 11.84VGGVCQSIVTAHH208 pKa = 6.52NEE210 pKa = 3.77FLHH213 pKa = 7.04AGLSDD218 pKa = 4.31DD219 pKa = 5.26RR220 pKa = 11.84GIPQAKK226 pKa = 8.98KK227 pKa = 9.96RR228 pKa = 11.84KK229 pKa = 9.03RR230 pKa = 11.84DD231 pKa = 3.53QGPHH235 pKa = 6.57KK236 pKa = 10.43GPPALPKK243 pKa = 10.53
Molecular weight: 26.51 kDa
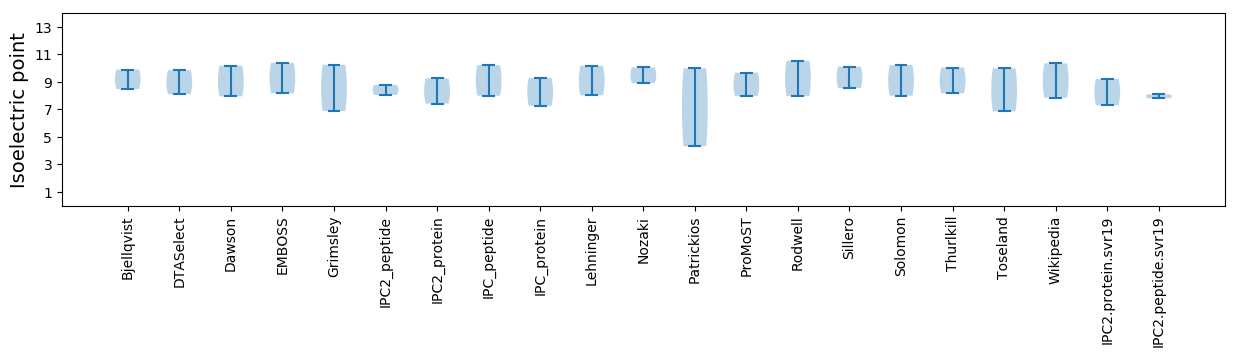
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHU0|A0A1L3KHU0_9VIRU Uncharacterized protein OS=Hubei mosquito virus 3 OX=1922927 PE=4 SV=1
MM1 pKa = 7.25VRR3 pKa = 11.84RR4 pKa = 11.84AYY6 pKa = 9.93DD7 pKa = 3.56LKK9 pKa = 11.09RR10 pKa = 11.84PLPYY14 pKa = 10.44SQVEE18 pKa = 3.95AAKK21 pKa = 10.15PCAHH25 pKa = 6.22QVKK28 pKa = 8.01TWKK31 pKa = 10.45EE32 pKa = 3.78FTNFLLDD39 pKa = 3.89LVAHH43 pKa = 6.72GKK45 pKa = 9.33SALVRR50 pKa = 11.84RR51 pKa = 11.84PGSTPFTRR59 pKa = 11.84SFTSFIVSRR68 pKa = 11.84CAWSRR73 pKa = 11.84GRR75 pKa = 11.84GKK77 pKa = 9.2WASIAFSLGQLKK89 pKa = 10.13RR90 pKa = 11.84CFPALPDD97 pKa = 3.43EE98 pKa = 4.55MRR100 pKa = 11.84RR101 pKa = 11.84EE102 pKa = 4.19ALDD105 pKa = 3.16KK106 pKa = 10.94HH107 pKa = 6.13QEE109 pKa = 4.18LLGSAAEE116 pKa = 4.29PFVEE120 pKa = 5.15FPIVDD125 pKa = 5.01RR126 pKa = 11.84IIQQYY131 pKa = 9.58FPVNWARR138 pKa = 11.84KK139 pKa = 7.23TSVKK143 pKa = 10.32VYY145 pKa = 10.62NAIPNTACIEE155 pKa = 4.37GGSCRR160 pKa = 11.84DD161 pKa = 3.69YY162 pKa = 11.21LASTGPKK169 pKa = 8.65PARR172 pKa = 11.84GCTPIPSSGQILAKK186 pKa = 9.95RR187 pKa = 11.84RR188 pKa = 11.84YY189 pKa = 9.2EE190 pKa = 4.31SIFDD194 pKa = 3.75KK195 pKa = 11.19LYY197 pKa = 8.86MQRR200 pKa = 11.84LQPLPTLKK208 pKa = 10.64AVAVPDD214 pKa = 3.71AGKK217 pKa = 10.25FRR219 pKa = 11.84IITIDD224 pKa = 3.58SVSAKK229 pKa = 9.98CLQPVQRR236 pKa = 11.84ALLSHH241 pKa = 7.09LSRR244 pKa = 11.84FPEE247 pKa = 4.41FEE249 pKa = 4.05FCTSTFQGEE258 pKa = 4.59LPPKK262 pKa = 7.45WGKK265 pKa = 7.05MAKK268 pKa = 9.91GEE270 pKa = 4.14KK271 pKa = 9.82LLSVDD276 pKa = 3.99YY277 pKa = 11.53SNATDD282 pKa = 3.95GVDD285 pKa = 3.19GSFMSEE291 pKa = 3.69LMDD294 pKa = 5.59RR295 pKa = 11.84IIDD298 pKa = 3.53RR299 pKa = 11.84SGSFEE304 pKa = 4.22LLALRR309 pKa = 11.84EE310 pKa = 4.02QAVQEE315 pKa = 4.3TTLGRR320 pKa = 11.84RR321 pKa = 11.84ISNDD325 pKa = 2.83LCSFWQRR332 pKa = 11.84RR333 pKa = 11.84GTLMGSLVSFPLLCLWNASIIRR355 pKa = 11.84EE356 pKa = 3.99AGMKK360 pKa = 10.2KK361 pKa = 10.54FIVCGDD367 pKa = 3.69DD368 pKa = 3.11ALAYY372 pKa = 7.74ATNTQKK378 pKa = 10.57KK379 pKa = 8.18RR380 pKa = 11.84WADD383 pKa = 3.45YY384 pKa = 9.88SAKK387 pKa = 10.34LGLVQSPGKK396 pKa = 8.36TFHH399 pKa = 6.22SRR401 pKa = 11.84KK402 pKa = 9.86FGTFCSKK409 pKa = 10.59VVGTEE414 pKa = 3.57HH415 pKa = 7.03LNAGRR420 pKa = 11.84ILAPMSGEE428 pKa = 4.01NYY430 pKa = 10.12NRR432 pKa = 11.84TPKK435 pKa = 10.06QFRR438 pKa = 11.84HH439 pKa = 5.26IYY441 pKa = 9.83KK442 pKa = 9.85RR443 pKa = 11.84YY444 pKa = 8.5CFYY447 pKa = 10.59PHH449 pKa = 6.93GEE451 pKa = 3.94FRR453 pKa = 11.84SLYY456 pKa = 9.11GPRR459 pKa = 11.84EE460 pKa = 3.74IGGLGGEE467 pKa = 4.36VDD469 pKa = 5.37DD470 pKa = 5.15IDD472 pKa = 4.16VPSRR476 pKa = 11.84RR477 pKa = 11.84ALRR480 pKa = 11.84KK481 pKa = 9.37AQAHH485 pKa = 6.83LARR488 pKa = 11.84QGTKK492 pKa = 9.98YY493 pKa = 10.1QLGMLDD499 pKa = 3.64MQDD502 pKa = 3.55EE503 pKa = 4.44NHH505 pKa = 6.5AGIYY509 pKa = 5.33TTAWSACDD517 pKa = 3.54PTPQRR522 pKa = 11.84LTPRR526 pKa = 11.84LRR528 pKa = 11.84KK529 pKa = 9.97GGSLDD534 pKa = 3.58YY535 pKa = 9.46RR536 pKa = 11.84TKK538 pKa = 9.95PIRR541 pKa = 11.84WFTRR545 pKa = 11.84WDD547 pKa = 3.39RR548 pKa = 11.84KK549 pKa = 9.53SRR551 pKa = 11.84SRR553 pKa = 11.84RR554 pKa = 11.84FF555 pKa = 3.14
MM1 pKa = 7.25VRR3 pKa = 11.84RR4 pKa = 11.84AYY6 pKa = 9.93DD7 pKa = 3.56LKK9 pKa = 11.09RR10 pKa = 11.84PLPYY14 pKa = 10.44SQVEE18 pKa = 3.95AAKK21 pKa = 10.15PCAHH25 pKa = 6.22QVKK28 pKa = 8.01TWKK31 pKa = 10.45EE32 pKa = 3.78FTNFLLDD39 pKa = 3.89LVAHH43 pKa = 6.72GKK45 pKa = 9.33SALVRR50 pKa = 11.84RR51 pKa = 11.84PGSTPFTRR59 pKa = 11.84SFTSFIVSRR68 pKa = 11.84CAWSRR73 pKa = 11.84GRR75 pKa = 11.84GKK77 pKa = 9.2WASIAFSLGQLKK89 pKa = 10.13RR90 pKa = 11.84CFPALPDD97 pKa = 3.43EE98 pKa = 4.55MRR100 pKa = 11.84RR101 pKa = 11.84EE102 pKa = 4.19ALDD105 pKa = 3.16KK106 pKa = 10.94HH107 pKa = 6.13QEE109 pKa = 4.18LLGSAAEE116 pKa = 4.29PFVEE120 pKa = 5.15FPIVDD125 pKa = 5.01RR126 pKa = 11.84IIQQYY131 pKa = 9.58FPVNWARR138 pKa = 11.84KK139 pKa = 7.23TSVKK143 pKa = 10.32VYY145 pKa = 10.62NAIPNTACIEE155 pKa = 4.37GGSCRR160 pKa = 11.84DD161 pKa = 3.69YY162 pKa = 11.21LASTGPKK169 pKa = 8.65PARR172 pKa = 11.84GCTPIPSSGQILAKK186 pKa = 9.95RR187 pKa = 11.84RR188 pKa = 11.84YY189 pKa = 9.2EE190 pKa = 4.31SIFDD194 pKa = 3.75KK195 pKa = 11.19LYY197 pKa = 8.86MQRR200 pKa = 11.84LQPLPTLKK208 pKa = 10.64AVAVPDD214 pKa = 3.71AGKK217 pKa = 10.25FRR219 pKa = 11.84IITIDD224 pKa = 3.58SVSAKK229 pKa = 9.98CLQPVQRR236 pKa = 11.84ALLSHH241 pKa = 7.09LSRR244 pKa = 11.84FPEE247 pKa = 4.41FEE249 pKa = 4.05FCTSTFQGEE258 pKa = 4.59LPPKK262 pKa = 7.45WGKK265 pKa = 7.05MAKK268 pKa = 9.91GEE270 pKa = 4.14KK271 pKa = 9.82LLSVDD276 pKa = 3.99YY277 pKa = 11.53SNATDD282 pKa = 3.95GVDD285 pKa = 3.19GSFMSEE291 pKa = 3.69LMDD294 pKa = 5.59RR295 pKa = 11.84IIDD298 pKa = 3.53RR299 pKa = 11.84SGSFEE304 pKa = 4.22LLALRR309 pKa = 11.84EE310 pKa = 4.02QAVQEE315 pKa = 4.3TTLGRR320 pKa = 11.84RR321 pKa = 11.84ISNDD325 pKa = 2.83LCSFWQRR332 pKa = 11.84RR333 pKa = 11.84GTLMGSLVSFPLLCLWNASIIRR355 pKa = 11.84EE356 pKa = 3.99AGMKK360 pKa = 10.2KK361 pKa = 10.54FIVCGDD367 pKa = 3.69DD368 pKa = 3.11ALAYY372 pKa = 7.74ATNTQKK378 pKa = 10.57KK379 pKa = 8.18RR380 pKa = 11.84WADD383 pKa = 3.45YY384 pKa = 9.88SAKK387 pKa = 10.34LGLVQSPGKK396 pKa = 8.36TFHH399 pKa = 6.22SRR401 pKa = 11.84KK402 pKa = 9.86FGTFCSKK409 pKa = 10.59VVGTEE414 pKa = 3.57HH415 pKa = 7.03LNAGRR420 pKa = 11.84ILAPMSGEE428 pKa = 4.01NYY430 pKa = 10.12NRR432 pKa = 11.84TPKK435 pKa = 10.06QFRR438 pKa = 11.84HH439 pKa = 5.26IYY441 pKa = 9.83KK442 pKa = 9.85RR443 pKa = 11.84YY444 pKa = 8.5CFYY447 pKa = 10.59PHH449 pKa = 6.93GEE451 pKa = 3.94FRR453 pKa = 11.84SLYY456 pKa = 9.11GPRR459 pKa = 11.84EE460 pKa = 3.74IGGLGGEE467 pKa = 4.36VDD469 pKa = 5.37DD470 pKa = 5.15IDD472 pKa = 4.16VPSRR476 pKa = 11.84RR477 pKa = 11.84ALRR480 pKa = 11.84KK481 pKa = 9.37AQAHH485 pKa = 6.83LARR488 pKa = 11.84QGTKK492 pKa = 9.98YY493 pKa = 10.1QLGMLDD499 pKa = 3.64MQDD502 pKa = 3.55EE503 pKa = 4.44NHH505 pKa = 6.5AGIYY509 pKa = 5.33TTAWSACDD517 pKa = 3.54PTPQRR522 pKa = 11.84LTPRR526 pKa = 11.84LRR528 pKa = 11.84KK529 pKa = 9.97GGSLDD534 pKa = 3.58YY535 pKa = 9.46RR536 pKa = 11.84TKK538 pKa = 9.95PIRR541 pKa = 11.84WFTRR545 pKa = 11.84WDD547 pKa = 3.39RR548 pKa = 11.84KK549 pKa = 9.53SRR551 pKa = 11.84SRR553 pKa = 11.84RR554 pKa = 11.84FF555 pKa = 3.14
Molecular weight: 62.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
798 |
243 |
555 |
399.0 |
44.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.268 ± 0.746 | 3.258 ± 0.832 |
4.386 ± 0.338 | 4.637 ± 0.353 |
4.887 ± 0.382 | 7.769 ± 0.636 |
2.256 ± 0.513 | 4.511 ± 0.008 |
5.263 ± 1.179 | 8.897 ± 0.126 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.13 ± 0.168 | 1.88 ± 0.319 |
7.018 ± 1.212 | 4.386 ± 0.273 |
8.897 ± 0.534 | 7.895 ± 0.166 |
5.138 ± 0.303 | 5.388 ± 0.999 |
1.629 ± 0.399 | 2.506 ± 0.833 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
