
Botrytis cinerea mitovirus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S4GA85|A0A0S4GA85_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 4 OX=1629667 GN=RdRp PE=4 SV=1
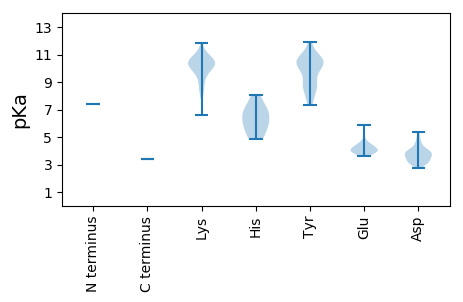
MM1 pKa = 7.42SNKK4 pKa = 9.58IINIIIIRR12 pKa = 11.84IIRR15 pKa = 11.84LCFNYY20 pKa = 10.38NEE22 pKa = 4.58STWEE26 pKa = 3.63INEE29 pKa = 3.97FLNIFNNMRR38 pKa = 11.84KK39 pKa = 9.71KK40 pKa = 10.69SGLKK44 pKa = 8.67YY45 pKa = 8.58TIKK48 pKa = 10.49YY49 pKa = 7.87YY50 pKa = 10.74KK51 pKa = 10.03AVKK54 pKa = 9.98LHH56 pKa = 4.82ITRR59 pKa = 11.84YY60 pKa = 9.05ICGSPLLSNKK70 pKa = 10.39DD71 pKa = 3.51GVSVDD76 pKa = 3.21ASGWPTKK83 pKa = 10.53FIFLKK88 pKa = 10.71KK89 pKa = 10.42FIKK92 pKa = 9.27TRR94 pKa = 11.84QGLRR98 pKa = 11.84ILLTLLSFTRR108 pKa = 11.84TVVPTKK114 pKa = 10.4QEE116 pKa = 3.87EE117 pKa = 4.39LKK119 pKa = 10.65IKK121 pKa = 10.05PDD123 pKa = 3.26YY124 pKa = 10.33STIDD128 pKa = 3.11KK129 pKa = 10.84LYY131 pKa = 8.7TGKK134 pKa = 10.51NYY136 pKa = 8.37TIPAWFIKK144 pKa = 10.26SWINKK149 pKa = 8.85HH150 pKa = 4.87NLKK153 pKa = 10.74AKK155 pKa = 9.84IPTYY159 pKa = 9.91TKK161 pKa = 8.87EE162 pKa = 3.73DD163 pKa = 3.86HH164 pKa = 5.92YY165 pKa = 11.88VSMKK169 pKa = 10.46GSPNGPATYY178 pKa = 10.43SSLWSILSLSYY189 pKa = 10.28PQLQNISTMLGDD201 pKa = 3.77YY202 pKa = 10.32RR203 pKa = 11.84DD204 pKa = 3.95EE205 pKa = 4.05FFKK208 pKa = 10.66FYY210 pKa = 8.96KK211 pKa = 9.24TAWEE215 pKa = 4.19NNFGNDD221 pKa = 3.96SSIKK225 pKa = 8.29SSKK228 pKa = 6.63WTGYY232 pKa = 7.77TGKK235 pKa = 10.61LSIVKK240 pKa = 10.24DD241 pKa = 3.69PEE243 pKa = 3.88LKK245 pKa = 10.27RR246 pKa = 11.84RR247 pKa = 11.84VIAMVDD253 pKa = 3.16YY254 pKa = 10.83HH255 pKa = 6.82SQFTLKK261 pKa = 10.17PIHH264 pKa = 6.83EE265 pKa = 4.35MLLNKK270 pKa = 9.89LRR272 pKa = 11.84TLKK275 pKa = 10.44CDD277 pKa = 3.05RR278 pKa = 11.84TFTQDD283 pKa = 4.13PKK285 pKa = 10.98HH286 pKa = 5.65SWYY289 pKa = 10.96VNNEE293 pKa = 3.79KK294 pKa = 10.47FFSLDD299 pKa = 3.02LSAATDD305 pKa = 3.96RR306 pKa = 11.84FPLQLQKK313 pKa = 11.35KK314 pKa = 8.25LLSYY318 pKa = 10.27IYY320 pKa = 9.76EE321 pKa = 4.29NKK323 pKa = 10.31DD324 pKa = 3.3FCDD327 pKa = 3.16AWADD331 pKa = 3.81LLTSRR336 pKa = 11.84VYY338 pKa = 10.45IDD340 pKa = 4.66SDD342 pKa = 3.94GLQHH346 pKa = 7.08KK347 pKa = 9.93YY348 pKa = 10.81NVGQPMGAYY357 pKa = 9.89SSWAAFTITHH367 pKa = 6.31HH368 pKa = 7.05LVVAWAAHH376 pKa = 5.7LCGEE380 pKa = 4.62YY381 pKa = 10.72NFSQYY386 pKa = 10.7IILGDD391 pKa = 4.59DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.9YY404 pKa = 7.31ITLMTRR410 pKa = 11.84LGVEE414 pKa = 3.87ISLHH418 pKa = 5.06KK419 pKa = 9.43THH421 pKa = 6.28VSKK424 pKa = 10.14DD425 pKa = 3.14TYY427 pKa = 10.9EE428 pKa = 3.97FAKK431 pKa = 10.31RR432 pKa = 11.84WIKK435 pKa = 10.86DD436 pKa = 3.54GIEE439 pKa = 3.85VSGIPLKK446 pKa = 11.08GILNQWKK453 pKa = 9.43FPGVVYY459 pKa = 7.53TTLEE463 pKa = 4.01SFFDD467 pKa = 3.93KK468 pKa = 11.24NPIQPKK474 pKa = 10.21SLIDD478 pKa = 4.57LICGLYY484 pKa = 10.57KK485 pKa = 10.53NLPLGKK491 pKa = 9.52RR492 pKa = 11.84RR493 pKa = 11.84MSYY496 pKa = 9.03NQIYY500 pKa = 9.96KK501 pKa = 10.66LLYY504 pKa = 9.8DD505 pKa = 3.43YY506 pKa = 10.54HH507 pKa = 7.85HH508 pKa = 7.3AMRR511 pKa = 11.84YY512 pKa = 8.49SLNKK516 pKa = 8.22ITYY519 pKa = 8.87DD520 pKa = 3.26EE521 pKa = 4.26LRR523 pKa = 11.84AYY525 pKa = 10.44LCSKK529 pKa = 10.24CKK531 pKa = 9.95EE532 pKa = 4.09DD533 pKa = 5.1SFVLPYY539 pKa = 10.74EE540 pKa = 4.66SISLHH545 pKa = 5.51FMKK548 pKa = 10.85LLLSGGMVSEE558 pKa = 4.35AEE560 pKa = 4.12KK561 pKa = 10.51VSRR564 pKa = 11.84FILSEE569 pKa = 3.8YY570 pKa = 10.12TKK572 pKa = 10.34IEE574 pKa = 4.08NKK576 pKa = 10.28FKK578 pKa = 11.0DD579 pKa = 3.62SYY581 pKa = 11.65SDD583 pKa = 4.09LNILSGYY590 pKa = 9.03PLLNGYY596 pKa = 8.75YY597 pKa = 10.24NHH599 pKa = 6.59LQSMQGKK606 pKa = 8.93ILDD609 pKa = 3.83WEE611 pKa = 4.23KK612 pKa = 11.35DD613 pKa = 3.63PNVTLVDD620 pKa = 3.62SALSLRR626 pKa = 11.84IEE628 pKa = 4.26KK629 pKa = 9.96FDD631 pKa = 5.36KK632 pKa = 10.28ISSMNRR638 pKa = 11.84DD639 pKa = 2.71KK640 pKa = 11.04SVRR643 pKa = 11.84VSTLSGLWKK652 pKa = 9.65TSMKK656 pKa = 10.2RR657 pKa = 11.84LWVEE661 pKa = 4.06RR662 pKa = 11.84IEE664 pKa = 5.88DD665 pKa = 3.75DD666 pKa = 3.74FEE668 pKa = 5.34YY669 pKa = 8.29MTFFSRR675 pKa = 11.84INKK678 pKa = 8.66DD679 pKa = 3.01QHH681 pKa = 8.09DD682 pKa = 4.3SMLPVHH688 pKa = 6.14GWEE691 pKa = 4.4GVLDD695 pKa = 3.97NNIQFTLNQLKK706 pKa = 9.83PLISGTIVKK715 pKa = 9.71VEE717 pKa = 4.01KK718 pKa = 10.92NSWEE722 pKa = 3.96NLDD725 pKa = 3.43WGDD728 pKa = 4.03FKK730 pKa = 11.81VV731 pKa = 3.4
MM1 pKa = 7.42SNKK4 pKa = 9.58IINIIIIRR12 pKa = 11.84IIRR15 pKa = 11.84LCFNYY20 pKa = 10.38NEE22 pKa = 4.58STWEE26 pKa = 3.63INEE29 pKa = 3.97FLNIFNNMRR38 pKa = 11.84KK39 pKa = 9.71KK40 pKa = 10.69SGLKK44 pKa = 8.67YY45 pKa = 8.58TIKK48 pKa = 10.49YY49 pKa = 7.87YY50 pKa = 10.74KK51 pKa = 10.03AVKK54 pKa = 9.98LHH56 pKa = 4.82ITRR59 pKa = 11.84YY60 pKa = 9.05ICGSPLLSNKK70 pKa = 10.39DD71 pKa = 3.51GVSVDD76 pKa = 3.21ASGWPTKK83 pKa = 10.53FIFLKK88 pKa = 10.71KK89 pKa = 10.42FIKK92 pKa = 9.27TRR94 pKa = 11.84QGLRR98 pKa = 11.84ILLTLLSFTRR108 pKa = 11.84TVVPTKK114 pKa = 10.4QEE116 pKa = 3.87EE117 pKa = 4.39LKK119 pKa = 10.65IKK121 pKa = 10.05PDD123 pKa = 3.26YY124 pKa = 10.33STIDD128 pKa = 3.11KK129 pKa = 10.84LYY131 pKa = 8.7TGKK134 pKa = 10.51NYY136 pKa = 8.37TIPAWFIKK144 pKa = 10.26SWINKK149 pKa = 8.85HH150 pKa = 4.87NLKK153 pKa = 10.74AKK155 pKa = 9.84IPTYY159 pKa = 9.91TKK161 pKa = 8.87EE162 pKa = 3.73DD163 pKa = 3.86HH164 pKa = 5.92YY165 pKa = 11.88VSMKK169 pKa = 10.46GSPNGPATYY178 pKa = 10.43SSLWSILSLSYY189 pKa = 10.28PQLQNISTMLGDD201 pKa = 3.77YY202 pKa = 10.32RR203 pKa = 11.84DD204 pKa = 3.95EE205 pKa = 4.05FFKK208 pKa = 10.66FYY210 pKa = 8.96KK211 pKa = 9.24TAWEE215 pKa = 4.19NNFGNDD221 pKa = 3.96SSIKK225 pKa = 8.29SSKK228 pKa = 6.63WTGYY232 pKa = 7.77TGKK235 pKa = 10.61LSIVKK240 pKa = 10.24DD241 pKa = 3.69PEE243 pKa = 3.88LKK245 pKa = 10.27RR246 pKa = 11.84RR247 pKa = 11.84VIAMVDD253 pKa = 3.16YY254 pKa = 10.83HH255 pKa = 6.82SQFTLKK261 pKa = 10.17PIHH264 pKa = 6.83EE265 pKa = 4.35MLLNKK270 pKa = 9.89LRR272 pKa = 11.84TLKK275 pKa = 10.44CDD277 pKa = 3.05RR278 pKa = 11.84TFTQDD283 pKa = 4.13PKK285 pKa = 10.98HH286 pKa = 5.65SWYY289 pKa = 10.96VNNEE293 pKa = 3.79KK294 pKa = 10.47FFSLDD299 pKa = 3.02LSAATDD305 pKa = 3.96RR306 pKa = 11.84FPLQLQKK313 pKa = 11.35KK314 pKa = 8.25LLSYY318 pKa = 10.27IYY320 pKa = 9.76EE321 pKa = 4.29NKK323 pKa = 10.31DD324 pKa = 3.3FCDD327 pKa = 3.16AWADD331 pKa = 3.81LLTSRR336 pKa = 11.84VYY338 pKa = 10.45IDD340 pKa = 4.66SDD342 pKa = 3.94GLQHH346 pKa = 7.08KK347 pKa = 9.93YY348 pKa = 10.81NVGQPMGAYY357 pKa = 9.89SSWAAFTITHH367 pKa = 6.31HH368 pKa = 7.05LVVAWAAHH376 pKa = 5.7LCGEE380 pKa = 4.62YY381 pKa = 10.72NFSQYY386 pKa = 10.7IILGDD391 pKa = 4.59DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.9YY404 pKa = 7.31ITLMTRR410 pKa = 11.84LGVEE414 pKa = 3.87ISLHH418 pKa = 5.06KK419 pKa = 9.43THH421 pKa = 6.28VSKK424 pKa = 10.14DD425 pKa = 3.14TYY427 pKa = 10.9EE428 pKa = 3.97FAKK431 pKa = 10.31RR432 pKa = 11.84WIKK435 pKa = 10.86DD436 pKa = 3.54GIEE439 pKa = 3.85VSGIPLKK446 pKa = 11.08GILNQWKK453 pKa = 9.43FPGVVYY459 pKa = 7.53TTLEE463 pKa = 4.01SFFDD467 pKa = 3.93KK468 pKa = 11.24NPIQPKK474 pKa = 10.21SLIDD478 pKa = 4.57LICGLYY484 pKa = 10.57KK485 pKa = 10.53NLPLGKK491 pKa = 9.52RR492 pKa = 11.84RR493 pKa = 11.84MSYY496 pKa = 9.03NQIYY500 pKa = 9.96KK501 pKa = 10.66LLYY504 pKa = 9.8DD505 pKa = 3.43YY506 pKa = 10.54HH507 pKa = 7.85HH508 pKa = 7.3AMRR511 pKa = 11.84YY512 pKa = 8.49SLNKK516 pKa = 8.22ITYY519 pKa = 8.87DD520 pKa = 3.26EE521 pKa = 4.26LRR523 pKa = 11.84AYY525 pKa = 10.44LCSKK529 pKa = 10.24CKK531 pKa = 9.95EE532 pKa = 4.09DD533 pKa = 5.1SFVLPYY539 pKa = 10.74EE540 pKa = 4.66SISLHH545 pKa = 5.51FMKK548 pKa = 10.85LLLSGGMVSEE558 pKa = 4.35AEE560 pKa = 4.12KK561 pKa = 10.51VSRR564 pKa = 11.84FILSEE569 pKa = 3.8YY570 pKa = 10.12TKK572 pKa = 10.34IEE574 pKa = 4.08NKK576 pKa = 10.28FKK578 pKa = 11.0DD579 pKa = 3.62SYY581 pKa = 11.65SDD583 pKa = 4.09LNILSGYY590 pKa = 9.03PLLNGYY596 pKa = 8.75YY597 pKa = 10.24NHH599 pKa = 6.59LQSMQGKK606 pKa = 8.93ILDD609 pKa = 3.83WEE611 pKa = 4.23KK612 pKa = 11.35DD613 pKa = 3.63PNVTLVDD620 pKa = 3.62SALSLRR626 pKa = 11.84IEE628 pKa = 4.26KK629 pKa = 9.96FDD631 pKa = 5.36KK632 pKa = 10.28ISSMNRR638 pKa = 11.84DD639 pKa = 2.71KK640 pKa = 11.04SVRR643 pKa = 11.84VSTLSGLWKK652 pKa = 9.65TSMKK656 pKa = 10.2RR657 pKa = 11.84LWVEE661 pKa = 4.06RR662 pKa = 11.84IEE664 pKa = 5.88DD665 pKa = 3.75DD666 pKa = 3.74FEE668 pKa = 5.34YY669 pKa = 8.29MTFFSRR675 pKa = 11.84INKK678 pKa = 8.66DD679 pKa = 3.01QHH681 pKa = 8.09DD682 pKa = 4.3SMLPVHH688 pKa = 6.14GWEE691 pKa = 4.4GVLDD695 pKa = 3.97NNIQFTLNQLKK706 pKa = 9.83PLISGTIVKK715 pKa = 9.71VEE717 pKa = 4.01KK718 pKa = 10.92NSWEE722 pKa = 3.96NLDD725 pKa = 3.43WGDD728 pKa = 4.03FKK730 pKa = 11.81VV731 pKa = 3.4
Molecular weight: 85.31 kDa
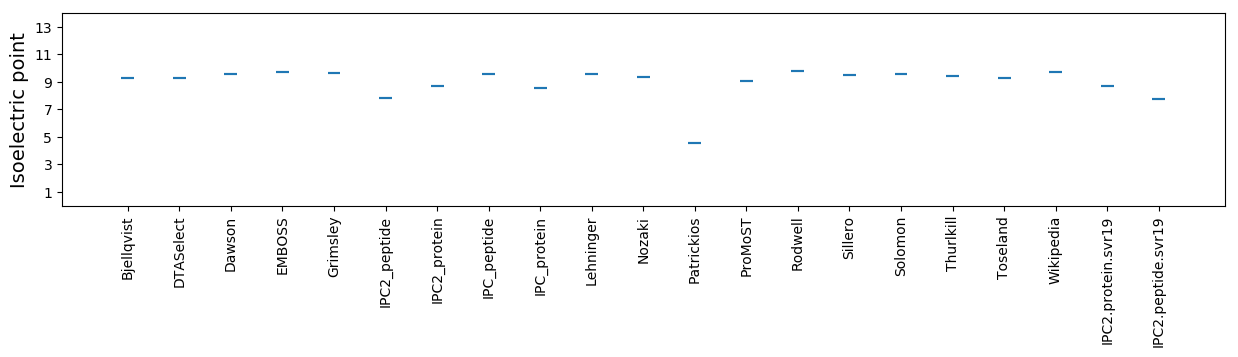
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S4GA85|A0A0S4GA85_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 4 OX=1629667 GN=RdRp PE=4 SV=1
MM1 pKa = 7.42SNKK4 pKa = 9.58IINIIIIRR12 pKa = 11.84IIRR15 pKa = 11.84LCFNYY20 pKa = 10.38NEE22 pKa = 4.58STWEE26 pKa = 3.63INEE29 pKa = 3.97FLNIFNNMRR38 pKa = 11.84KK39 pKa = 9.71KK40 pKa = 10.69SGLKK44 pKa = 8.67YY45 pKa = 8.58TIKK48 pKa = 10.49YY49 pKa = 7.87YY50 pKa = 10.74KK51 pKa = 10.03AVKK54 pKa = 9.98LHH56 pKa = 4.82ITRR59 pKa = 11.84YY60 pKa = 9.05ICGSPLLSNKK70 pKa = 10.39DD71 pKa = 3.51GVSVDD76 pKa = 3.21ASGWPTKK83 pKa = 10.53FIFLKK88 pKa = 10.71KK89 pKa = 10.42FIKK92 pKa = 9.27TRR94 pKa = 11.84QGLRR98 pKa = 11.84ILLTLLSFTRR108 pKa = 11.84TVVPTKK114 pKa = 10.4QEE116 pKa = 3.87EE117 pKa = 4.39LKK119 pKa = 10.65IKK121 pKa = 10.05PDD123 pKa = 3.26YY124 pKa = 10.33STIDD128 pKa = 3.11KK129 pKa = 10.84LYY131 pKa = 8.7TGKK134 pKa = 10.51NYY136 pKa = 8.37TIPAWFIKK144 pKa = 10.26SWINKK149 pKa = 8.85HH150 pKa = 4.87NLKK153 pKa = 10.74AKK155 pKa = 9.84IPTYY159 pKa = 9.91TKK161 pKa = 8.87EE162 pKa = 3.73DD163 pKa = 3.86HH164 pKa = 5.92YY165 pKa = 11.88VSMKK169 pKa = 10.46GSPNGPATYY178 pKa = 10.43SSLWSILSLSYY189 pKa = 10.28PQLQNISTMLGDD201 pKa = 3.77YY202 pKa = 10.32RR203 pKa = 11.84DD204 pKa = 3.95EE205 pKa = 4.05FFKK208 pKa = 10.66FYY210 pKa = 8.96KK211 pKa = 9.24TAWEE215 pKa = 4.19NNFGNDD221 pKa = 3.96SSIKK225 pKa = 8.29SSKK228 pKa = 6.63WTGYY232 pKa = 7.77TGKK235 pKa = 10.61LSIVKK240 pKa = 10.24DD241 pKa = 3.69PEE243 pKa = 3.88LKK245 pKa = 10.27RR246 pKa = 11.84RR247 pKa = 11.84VIAMVDD253 pKa = 3.16YY254 pKa = 10.83HH255 pKa = 6.82SQFTLKK261 pKa = 10.17PIHH264 pKa = 6.83EE265 pKa = 4.35MLLNKK270 pKa = 9.89LRR272 pKa = 11.84TLKK275 pKa = 10.44CDD277 pKa = 3.05RR278 pKa = 11.84TFTQDD283 pKa = 4.13PKK285 pKa = 10.98HH286 pKa = 5.65SWYY289 pKa = 10.96VNNEE293 pKa = 3.79KK294 pKa = 10.47FFSLDD299 pKa = 3.02LSAATDD305 pKa = 3.96RR306 pKa = 11.84FPLQLQKK313 pKa = 11.35KK314 pKa = 8.25LLSYY318 pKa = 10.27IYY320 pKa = 9.76EE321 pKa = 4.29NKK323 pKa = 10.31DD324 pKa = 3.3FCDD327 pKa = 3.16AWADD331 pKa = 3.81LLTSRR336 pKa = 11.84VYY338 pKa = 10.45IDD340 pKa = 4.66SDD342 pKa = 3.94GLQHH346 pKa = 7.08KK347 pKa = 9.93YY348 pKa = 10.81NVGQPMGAYY357 pKa = 9.89SSWAAFTITHH367 pKa = 6.31HH368 pKa = 7.05LVVAWAAHH376 pKa = 5.7LCGEE380 pKa = 4.62YY381 pKa = 10.72NFSQYY386 pKa = 10.7IILGDD391 pKa = 4.59DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.9YY404 pKa = 7.31ITLMTRR410 pKa = 11.84LGVEE414 pKa = 3.87ISLHH418 pKa = 5.06KK419 pKa = 9.43THH421 pKa = 6.28VSKK424 pKa = 10.14DD425 pKa = 3.14TYY427 pKa = 10.9EE428 pKa = 3.97FAKK431 pKa = 10.31RR432 pKa = 11.84WIKK435 pKa = 10.86DD436 pKa = 3.54GIEE439 pKa = 3.85VSGIPLKK446 pKa = 11.08GILNQWKK453 pKa = 9.43FPGVVYY459 pKa = 7.53TTLEE463 pKa = 4.01SFFDD467 pKa = 3.93KK468 pKa = 11.24NPIQPKK474 pKa = 10.21SLIDD478 pKa = 4.57LICGLYY484 pKa = 10.57KK485 pKa = 10.53NLPLGKK491 pKa = 9.52RR492 pKa = 11.84RR493 pKa = 11.84MSYY496 pKa = 9.03NQIYY500 pKa = 9.96KK501 pKa = 10.66LLYY504 pKa = 9.8DD505 pKa = 3.43YY506 pKa = 10.54HH507 pKa = 7.85HH508 pKa = 7.3AMRR511 pKa = 11.84YY512 pKa = 8.49SLNKK516 pKa = 8.22ITYY519 pKa = 8.87DD520 pKa = 3.26EE521 pKa = 4.26LRR523 pKa = 11.84AYY525 pKa = 10.44LCSKK529 pKa = 10.24CKK531 pKa = 9.95EE532 pKa = 4.09DD533 pKa = 5.1SFVLPYY539 pKa = 10.74EE540 pKa = 4.66SISLHH545 pKa = 5.51FMKK548 pKa = 10.85LLLSGGMVSEE558 pKa = 4.35AEE560 pKa = 4.12KK561 pKa = 10.51VSRR564 pKa = 11.84FILSEE569 pKa = 3.8YY570 pKa = 10.12TKK572 pKa = 10.34IEE574 pKa = 4.08NKK576 pKa = 10.28FKK578 pKa = 11.0DD579 pKa = 3.62SYY581 pKa = 11.65SDD583 pKa = 4.09LNILSGYY590 pKa = 9.03PLLNGYY596 pKa = 8.75YY597 pKa = 10.24NHH599 pKa = 6.59LQSMQGKK606 pKa = 8.93ILDD609 pKa = 3.83WEE611 pKa = 4.23KK612 pKa = 11.35DD613 pKa = 3.63PNVTLVDD620 pKa = 3.62SALSLRR626 pKa = 11.84IEE628 pKa = 4.26KK629 pKa = 9.96FDD631 pKa = 5.36KK632 pKa = 10.28ISSMNRR638 pKa = 11.84DD639 pKa = 2.71KK640 pKa = 11.04SVRR643 pKa = 11.84VSTLSGLWKK652 pKa = 9.65TSMKK656 pKa = 10.2RR657 pKa = 11.84LWVEE661 pKa = 4.06RR662 pKa = 11.84IEE664 pKa = 5.88DD665 pKa = 3.75DD666 pKa = 3.74FEE668 pKa = 5.34YY669 pKa = 8.29MTFFSRR675 pKa = 11.84INKK678 pKa = 8.66DD679 pKa = 3.01QHH681 pKa = 8.09DD682 pKa = 4.3SMLPVHH688 pKa = 6.14GWEE691 pKa = 4.4GVLDD695 pKa = 3.97NNIQFTLNQLKK706 pKa = 9.83PLISGTIVKK715 pKa = 9.71VEE717 pKa = 4.01KK718 pKa = 10.92NSWEE722 pKa = 3.96NLDD725 pKa = 3.43WGDD728 pKa = 4.03FKK730 pKa = 11.81VV731 pKa = 3.4
MM1 pKa = 7.42SNKK4 pKa = 9.58IINIIIIRR12 pKa = 11.84IIRR15 pKa = 11.84LCFNYY20 pKa = 10.38NEE22 pKa = 4.58STWEE26 pKa = 3.63INEE29 pKa = 3.97FLNIFNNMRR38 pKa = 11.84KK39 pKa = 9.71KK40 pKa = 10.69SGLKK44 pKa = 8.67YY45 pKa = 8.58TIKK48 pKa = 10.49YY49 pKa = 7.87YY50 pKa = 10.74KK51 pKa = 10.03AVKK54 pKa = 9.98LHH56 pKa = 4.82ITRR59 pKa = 11.84YY60 pKa = 9.05ICGSPLLSNKK70 pKa = 10.39DD71 pKa = 3.51GVSVDD76 pKa = 3.21ASGWPTKK83 pKa = 10.53FIFLKK88 pKa = 10.71KK89 pKa = 10.42FIKK92 pKa = 9.27TRR94 pKa = 11.84QGLRR98 pKa = 11.84ILLTLLSFTRR108 pKa = 11.84TVVPTKK114 pKa = 10.4QEE116 pKa = 3.87EE117 pKa = 4.39LKK119 pKa = 10.65IKK121 pKa = 10.05PDD123 pKa = 3.26YY124 pKa = 10.33STIDD128 pKa = 3.11KK129 pKa = 10.84LYY131 pKa = 8.7TGKK134 pKa = 10.51NYY136 pKa = 8.37TIPAWFIKK144 pKa = 10.26SWINKK149 pKa = 8.85HH150 pKa = 4.87NLKK153 pKa = 10.74AKK155 pKa = 9.84IPTYY159 pKa = 9.91TKK161 pKa = 8.87EE162 pKa = 3.73DD163 pKa = 3.86HH164 pKa = 5.92YY165 pKa = 11.88VSMKK169 pKa = 10.46GSPNGPATYY178 pKa = 10.43SSLWSILSLSYY189 pKa = 10.28PQLQNISTMLGDD201 pKa = 3.77YY202 pKa = 10.32RR203 pKa = 11.84DD204 pKa = 3.95EE205 pKa = 4.05FFKK208 pKa = 10.66FYY210 pKa = 8.96KK211 pKa = 9.24TAWEE215 pKa = 4.19NNFGNDD221 pKa = 3.96SSIKK225 pKa = 8.29SSKK228 pKa = 6.63WTGYY232 pKa = 7.77TGKK235 pKa = 10.61LSIVKK240 pKa = 10.24DD241 pKa = 3.69PEE243 pKa = 3.88LKK245 pKa = 10.27RR246 pKa = 11.84RR247 pKa = 11.84VIAMVDD253 pKa = 3.16YY254 pKa = 10.83HH255 pKa = 6.82SQFTLKK261 pKa = 10.17PIHH264 pKa = 6.83EE265 pKa = 4.35MLLNKK270 pKa = 9.89LRR272 pKa = 11.84TLKK275 pKa = 10.44CDD277 pKa = 3.05RR278 pKa = 11.84TFTQDD283 pKa = 4.13PKK285 pKa = 10.98HH286 pKa = 5.65SWYY289 pKa = 10.96VNNEE293 pKa = 3.79KK294 pKa = 10.47FFSLDD299 pKa = 3.02LSAATDD305 pKa = 3.96RR306 pKa = 11.84FPLQLQKK313 pKa = 11.35KK314 pKa = 8.25LLSYY318 pKa = 10.27IYY320 pKa = 9.76EE321 pKa = 4.29NKK323 pKa = 10.31DD324 pKa = 3.3FCDD327 pKa = 3.16AWADD331 pKa = 3.81LLTSRR336 pKa = 11.84VYY338 pKa = 10.45IDD340 pKa = 4.66SDD342 pKa = 3.94GLQHH346 pKa = 7.08KK347 pKa = 9.93YY348 pKa = 10.81NVGQPMGAYY357 pKa = 9.89SSWAAFTITHH367 pKa = 6.31HH368 pKa = 7.05LVVAWAAHH376 pKa = 5.7LCGEE380 pKa = 4.62YY381 pKa = 10.72NFSQYY386 pKa = 10.7IILGDD391 pKa = 4.59DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.9YY404 pKa = 7.31ITLMTRR410 pKa = 11.84LGVEE414 pKa = 3.87ISLHH418 pKa = 5.06KK419 pKa = 9.43THH421 pKa = 6.28VSKK424 pKa = 10.14DD425 pKa = 3.14TYY427 pKa = 10.9EE428 pKa = 3.97FAKK431 pKa = 10.31RR432 pKa = 11.84WIKK435 pKa = 10.86DD436 pKa = 3.54GIEE439 pKa = 3.85VSGIPLKK446 pKa = 11.08GILNQWKK453 pKa = 9.43FPGVVYY459 pKa = 7.53TTLEE463 pKa = 4.01SFFDD467 pKa = 3.93KK468 pKa = 11.24NPIQPKK474 pKa = 10.21SLIDD478 pKa = 4.57LICGLYY484 pKa = 10.57KK485 pKa = 10.53NLPLGKK491 pKa = 9.52RR492 pKa = 11.84RR493 pKa = 11.84MSYY496 pKa = 9.03NQIYY500 pKa = 9.96KK501 pKa = 10.66LLYY504 pKa = 9.8DD505 pKa = 3.43YY506 pKa = 10.54HH507 pKa = 7.85HH508 pKa = 7.3AMRR511 pKa = 11.84YY512 pKa = 8.49SLNKK516 pKa = 8.22ITYY519 pKa = 8.87DD520 pKa = 3.26EE521 pKa = 4.26LRR523 pKa = 11.84AYY525 pKa = 10.44LCSKK529 pKa = 10.24CKK531 pKa = 9.95EE532 pKa = 4.09DD533 pKa = 5.1SFVLPYY539 pKa = 10.74EE540 pKa = 4.66SISLHH545 pKa = 5.51FMKK548 pKa = 10.85LLLSGGMVSEE558 pKa = 4.35AEE560 pKa = 4.12KK561 pKa = 10.51VSRR564 pKa = 11.84FILSEE569 pKa = 3.8YY570 pKa = 10.12TKK572 pKa = 10.34IEE574 pKa = 4.08NKK576 pKa = 10.28FKK578 pKa = 11.0DD579 pKa = 3.62SYY581 pKa = 11.65SDD583 pKa = 4.09LNILSGYY590 pKa = 9.03PLLNGYY596 pKa = 8.75YY597 pKa = 10.24NHH599 pKa = 6.59LQSMQGKK606 pKa = 8.93ILDD609 pKa = 3.83WEE611 pKa = 4.23KK612 pKa = 11.35DD613 pKa = 3.63PNVTLVDD620 pKa = 3.62SALSLRR626 pKa = 11.84IEE628 pKa = 4.26KK629 pKa = 9.96FDD631 pKa = 5.36KK632 pKa = 10.28ISSMNRR638 pKa = 11.84DD639 pKa = 2.71KK640 pKa = 11.04SVRR643 pKa = 11.84VSTLSGLWKK652 pKa = 9.65TSMKK656 pKa = 10.2RR657 pKa = 11.84LWVEE661 pKa = 4.06RR662 pKa = 11.84IEE664 pKa = 5.88DD665 pKa = 3.75DD666 pKa = 3.74FEE668 pKa = 5.34YY669 pKa = 8.29MTFFSRR675 pKa = 11.84INKK678 pKa = 8.66DD679 pKa = 3.01QHH681 pKa = 8.09DD682 pKa = 4.3SMLPVHH688 pKa = 6.14GWEE691 pKa = 4.4GVLDD695 pKa = 3.97NNIQFTLNQLKK706 pKa = 9.83PLISGTIVKK715 pKa = 9.71VEE717 pKa = 4.01KK718 pKa = 10.92NSWEE722 pKa = 3.96NLDD725 pKa = 3.43WGDD728 pKa = 4.03FKK730 pKa = 11.81VV731 pKa = 3.4
Molecular weight: 85.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
731 |
731 |
731 |
731.0 |
85.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.146 ± 0.0 | 1.094 ± 0.0 |
5.746 ± 0.0 | 4.378 ± 0.0 |
4.651 ± 0.0 | 4.651 ± 0.0 |
2.462 ± 0.0 | 7.934 ± 0.0 |
9.986 ± 0.0 | 10.807 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.0 | 5.882 ± 0.0 |
3.283 ± 0.0 | 2.599 ± 0.0 |
3.694 ± 0.0 | 8.755 ± 0.0 |
5.609 ± 0.0 | 4.651 ± 0.0 |
2.599 ± 0.0 | 5.746 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
