
Bacillus salarius
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
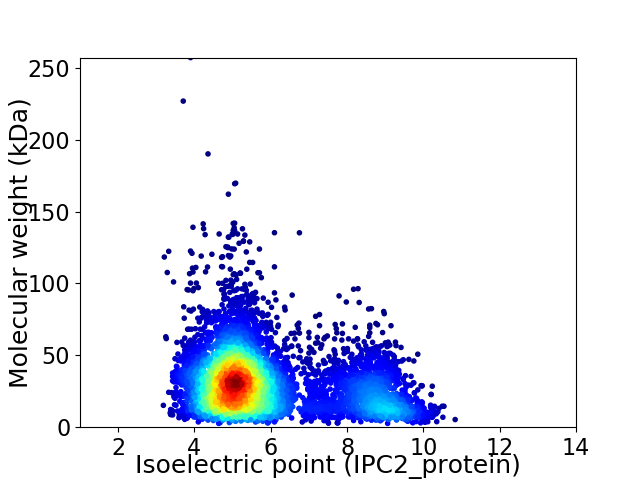
Virtual 2D-PAGE plot for 6761 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
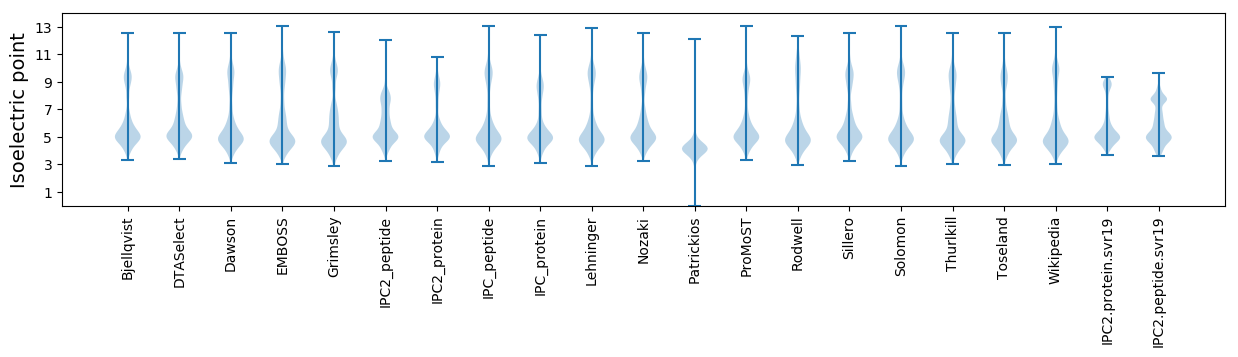
Protein with the lowest isoelectric point:
>tr|A0A3R9P520|A0A3R9P520_9BACI RNA methyltransferase OS=Bacillus salarius OX=284579 GN=D7Z54_13290 PE=4 SV=1
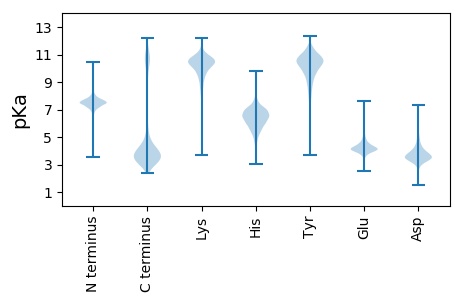
MM1 pKa = 7.21KK2 pKa = 10.36QLLQKK7 pKa = 10.55EE8 pKa = 4.51SGVTLIEE15 pKa = 4.02LLATIVISSIVIGLVTSVLVSSLNFNDD42 pKa = 3.58KK43 pKa = 9.03TQSHH47 pKa = 6.36INLRR51 pKa = 11.84QEE53 pKa = 3.74ANIIITEE60 pKa = 4.31LRR62 pKa = 11.84QQHH65 pKa = 5.28QEE67 pKa = 3.91GEE69 pKa = 4.36YY70 pKa = 9.14TLCPEE75 pKa = 4.99DD76 pKa = 4.09VFSSDD81 pKa = 3.18RR82 pKa = 11.84FRR84 pKa = 11.84AVQRR88 pKa = 11.84DD89 pKa = 3.36IRR91 pKa = 11.84NDD93 pKa = 2.71EE94 pKa = 4.31HH95 pKa = 7.01MITSCNTVDD104 pKa = 3.33SQFPLEE110 pKa = 4.18VQFTLEE116 pKa = 4.24DD117 pKa = 3.93DD118 pKa = 4.08EE119 pKa = 6.22NNDD122 pKa = 3.38FTIDD126 pKa = 3.66TIIEE130 pKa = 4.09GEE132 pKa = 4.17RR133 pKa = 11.84QNGDD137 pKa = 3.57TNVSIDD143 pKa = 3.74PPGDD147 pKa = 3.51EE148 pKa = 4.44SDD150 pKa = 3.88SFPTYY155 pKa = 11.02VEE157 pKa = 4.3DD158 pKa = 4.06EE159 pKa = 4.28NVFVYY164 pKa = 10.31GSQFTFQGSDD174 pKa = 3.3VNGPGASMVIKK185 pKa = 10.65GPLDD189 pKa = 3.19MSEE192 pKa = 4.52FNGGSKK198 pKa = 9.77TNVSNIYY205 pKa = 10.12VDD207 pKa = 3.99GPIDD211 pKa = 3.77FSGGGQDD218 pKa = 3.0LGSYY222 pKa = 9.09EE223 pKa = 4.5EE224 pKa = 4.77PGEE227 pKa = 3.77IHH229 pKa = 7.16INGDD233 pKa = 3.18FDD235 pKa = 4.07TGGGSHH241 pKa = 7.28NIYY244 pKa = 9.85GDD246 pKa = 3.66VYY248 pKa = 10.9VEE250 pKa = 4.16EE251 pKa = 5.24DD252 pKa = 3.46FHH254 pKa = 9.34LEE256 pKa = 3.69GANIYY261 pKa = 10.84GDD263 pKa = 3.77VYY265 pKa = 11.63VNGDD269 pKa = 3.49VTLSDD274 pKa = 4.23YY275 pKa = 11.63YY276 pKa = 11.36SIAKK280 pKa = 9.06NASIHH285 pKa = 4.76YY286 pKa = 7.83TGSLPYY292 pKa = 8.86PDD294 pKa = 4.0HH295 pKa = 7.25FEE297 pKa = 4.71RR298 pKa = 11.84SDD300 pKa = 3.42FDD302 pKa = 4.25SLVKK306 pKa = 10.51QEE308 pKa = 4.52SVPNAEE314 pKa = 4.84IPDD317 pKa = 3.76QEE319 pKa = 4.53VPSSKK324 pKa = 10.61SEE326 pKa = 3.28NWYY329 pKa = 10.72AEE331 pKa = 3.69NGYY334 pKa = 6.69TQEE337 pKa = 4.01IQEE340 pKa = 5.15DD341 pKa = 3.94GMKK344 pKa = 9.82IYY346 pKa = 10.88DD347 pKa = 3.8SDD349 pKa = 4.25VVIEE353 pKa = 4.46DD354 pKa = 4.03NVNGSYY360 pKa = 10.86QDD362 pKa = 3.37TFTDD366 pKa = 3.51SVVVSEE372 pKa = 4.4GDD374 pKa = 3.24ITISGGNLSMTGVLYY389 pKa = 10.78APNGEE394 pKa = 4.12ITFEE398 pKa = 4.15GASFEE403 pKa = 4.35GTVIAKK409 pKa = 10.44DD410 pKa = 3.63GFNVDD415 pKa = 3.86SGGTDD420 pKa = 2.71ITFVGVEE427 pKa = 4.01EE428 pKa = 4.71YY429 pKa = 10.71INNRR433 pKa = 11.84DD434 pKa = 3.84DD435 pKa = 3.81YY436 pKa = 11.32PFF438 pKa = 5.07
MM1 pKa = 7.21KK2 pKa = 10.36QLLQKK7 pKa = 10.55EE8 pKa = 4.51SGVTLIEE15 pKa = 4.02LLATIVISSIVIGLVTSVLVSSLNFNDD42 pKa = 3.58KK43 pKa = 9.03TQSHH47 pKa = 6.36INLRR51 pKa = 11.84QEE53 pKa = 3.74ANIIITEE60 pKa = 4.31LRR62 pKa = 11.84QQHH65 pKa = 5.28QEE67 pKa = 3.91GEE69 pKa = 4.36YY70 pKa = 9.14TLCPEE75 pKa = 4.99DD76 pKa = 4.09VFSSDD81 pKa = 3.18RR82 pKa = 11.84FRR84 pKa = 11.84AVQRR88 pKa = 11.84DD89 pKa = 3.36IRR91 pKa = 11.84NDD93 pKa = 2.71EE94 pKa = 4.31HH95 pKa = 7.01MITSCNTVDD104 pKa = 3.33SQFPLEE110 pKa = 4.18VQFTLEE116 pKa = 4.24DD117 pKa = 3.93DD118 pKa = 4.08EE119 pKa = 6.22NNDD122 pKa = 3.38FTIDD126 pKa = 3.66TIIEE130 pKa = 4.09GEE132 pKa = 4.17RR133 pKa = 11.84QNGDD137 pKa = 3.57TNVSIDD143 pKa = 3.74PPGDD147 pKa = 3.51EE148 pKa = 4.44SDD150 pKa = 3.88SFPTYY155 pKa = 11.02VEE157 pKa = 4.3DD158 pKa = 4.06EE159 pKa = 4.28NVFVYY164 pKa = 10.31GSQFTFQGSDD174 pKa = 3.3VNGPGASMVIKK185 pKa = 10.65GPLDD189 pKa = 3.19MSEE192 pKa = 4.52FNGGSKK198 pKa = 9.77TNVSNIYY205 pKa = 10.12VDD207 pKa = 3.99GPIDD211 pKa = 3.77FSGGGQDD218 pKa = 3.0LGSYY222 pKa = 9.09EE223 pKa = 4.5EE224 pKa = 4.77PGEE227 pKa = 3.77IHH229 pKa = 7.16INGDD233 pKa = 3.18FDD235 pKa = 4.07TGGGSHH241 pKa = 7.28NIYY244 pKa = 9.85GDD246 pKa = 3.66VYY248 pKa = 10.9VEE250 pKa = 4.16EE251 pKa = 5.24DD252 pKa = 3.46FHH254 pKa = 9.34LEE256 pKa = 3.69GANIYY261 pKa = 10.84GDD263 pKa = 3.77VYY265 pKa = 11.63VNGDD269 pKa = 3.49VTLSDD274 pKa = 4.23YY275 pKa = 11.63YY276 pKa = 11.36SIAKK280 pKa = 9.06NASIHH285 pKa = 4.76YY286 pKa = 7.83TGSLPYY292 pKa = 8.86PDD294 pKa = 4.0HH295 pKa = 7.25FEE297 pKa = 4.71RR298 pKa = 11.84SDD300 pKa = 3.42FDD302 pKa = 4.25SLVKK306 pKa = 10.51QEE308 pKa = 4.52SVPNAEE314 pKa = 4.84IPDD317 pKa = 3.76QEE319 pKa = 4.53VPSSKK324 pKa = 10.61SEE326 pKa = 3.28NWYY329 pKa = 10.72AEE331 pKa = 3.69NGYY334 pKa = 6.69TQEE337 pKa = 4.01IQEE340 pKa = 5.15DD341 pKa = 3.94GMKK344 pKa = 9.82IYY346 pKa = 10.88DD347 pKa = 3.8SDD349 pKa = 4.25VVIEE353 pKa = 4.46DD354 pKa = 4.03NVNGSYY360 pKa = 10.86QDD362 pKa = 3.37TFTDD366 pKa = 3.51SVVVSEE372 pKa = 4.4GDD374 pKa = 3.24ITISGGNLSMTGVLYY389 pKa = 10.78APNGEE394 pKa = 4.12ITFEE398 pKa = 4.15GASFEE403 pKa = 4.35GTVIAKK409 pKa = 10.44DD410 pKa = 3.63GFNVDD415 pKa = 3.86SGGTDD420 pKa = 2.71ITFVGVEE427 pKa = 4.01EE428 pKa = 4.71YY429 pKa = 10.71INNRR433 pKa = 11.84DD434 pKa = 3.84DD435 pKa = 3.81YY436 pKa = 11.32PFF438 pKa = 5.07
Molecular weight: 48.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3R9RCE8|A0A3R9RCE8_9BACI 3-isopropylmalate dehydrogenase OS=Bacillus salarius OX=284579 GN=leuB PE=3 SV=1
MM1 pKa = 7.67GKK3 pKa = 7.99PTFTPNNRR11 pKa = 11.84KK12 pKa = 9.22RR13 pKa = 11.84KK14 pKa = 7.99KK15 pKa = 8.43VHH17 pKa = 5.59GFRR20 pKa = 11.84EE21 pKa = 4.09RR22 pKa = 11.84MSTKK26 pKa = 10.29NGRR29 pKa = 11.84QVLKK33 pKa = 10.21SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.09GRR40 pKa = 11.84KK41 pKa = 9.06VISAA45 pKa = 4.05
MM1 pKa = 7.67GKK3 pKa = 7.99PTFTPNNRR11 pKa = 11.84KK12 pKa = 9.22RR13 pKa = 11.84KK14 pKa = 7.99KK15 pKa = 8.43VHH17 pKa = 5.59GFRR20 pKa = 11.84EE21 pKa = 4.09RR22 pKa = 11.84MSTKK26 pKa = 10.29NGRR29 pKa = 11.84QVLKK33 pKa = 10.21SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.09GRR40 pKa = 11.84KK41 pKa = 9.06VISAA45 pKa = 4.05
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1924827 |
24 |
2295 |
284.7 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.091 ± 0.034 | 0.675 ± 0.008 |
5.608 ± 0.029 | 8.26 ± 0.041 |
4.391 ± 0.024 | 6.932 ± 0.028 |
2.189 ± 0.015 | 7.207 ± 0.029 |
5.997 ± 0.029 | 9.242 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.966 ± 0.016 | 4.43 ± 0.02 |
3.651 ± 0.016 | 3.931 ± 0.023 |
4.063 ± 0.024 | 6.24 ± 0.024 |
5.636 ± 0.019 | 7.008 ± 0.024 |
1.087 ± 0.011 | 3.396 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
