
Candidatus Accumulibacter sp. BA-91
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Accumulibacter; unclassified Candidatus Accumulibacter
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
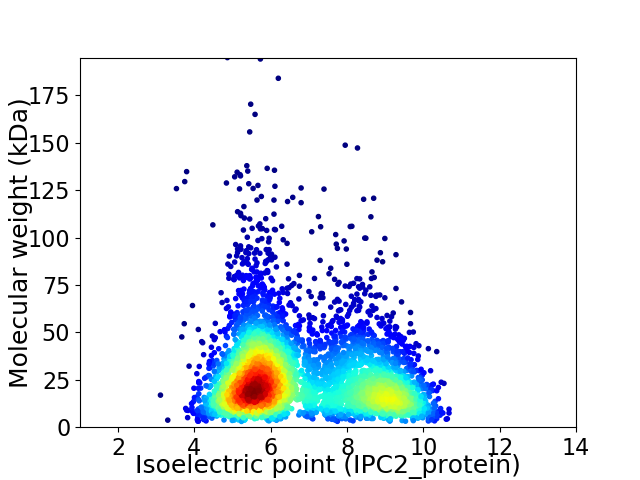
Virtual 2D-PAGE plot for 4449 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084Y6J2|A0A084Y6J2_9PROT 50S ribosomal protein L4 OS=Candidatus Accumulibacter sp. BA-91 OX=1454002 GN=rplD PE=3 SV=1
MM1 pKa = 7.57TIANLTLAQVLDD13 pKa = 3.8QLNSGRR19 pKa = 11.84SWSGASISYY28 pKa = 9.32SFPNTAPGLYY38 pKa = 9.92SQGEE42 pKa = 4.03AAAFRR47 pKa = 11.84AFDD50 pKa = 3.82ADD52 pKa = 3.54QRR54 pKa = 11.84SMMTLAMVTWDD65 pKa = 5.05DD66 pKa = 4.57LISPNFVQGTAGTTHH81 pKa = 7.43IEE83 pKa = 4.03FGYY86 pKa = 6.12TTSXIGFAHH95 pKa = 7.55AYY97 pKa = 8.33YY98 pKa = 10.17PEE100 pKa = 4.68IFGPRR105 pKa = 11.84YY106 pKa = 8.57AAPYY110 pKa = 8.72YY111 pKa = 10.46SSEE114 pKa = 4.02VMQADD119 pKa = 3.47WTGSDD124 pKa = 2.88GRR126 pKa = 11.84TYY128 pKa = 10.86SAQTPMLNDD137 pKa = 2.99VMAIQAIYY145 pKa = 9.65GASTSTRR152 pKa = 11.84LDD154 pKa = 3.11NTVYY158 pKa = 10.77GFGSNIVDD166 pKa = 3.79STRR169 pKa = 11.84AIYY172 pKa = 10.36DD173 pKa = 3.62FSLNAHH179 pKa = 7.61PILTLYY185 pKa = 10.61DD186 pKa = 3.54AGGLDD191 pKa = 3.97TLNLSGWSTPSRR203 pKa = 11.84IDD205 pKa = 3.02LHH207 pKa = 8.18AGAFSSGNDD216 pKa = 3.21MSNNIAIAYY225 pKa = 7.55NATIEE230 pKa = 4.05NAEE233 pKa = 4.18GGGGNDD239 pKa = 3.69VITGNDD245 pKa = 3.31TANLLRR251 pKa = 11.84GAAGNDD257 pKa = 3.36EE258 pKa = 5.1LYY260 pKa = 11.15GSAGDD265 pKa = 3.75DD266 pKa = 3.52TLIGGTGNDD275 pKa = 4.36DD276 pKa = 3.56LRR278 pKa = 11.84GGAGTDD284 pKa = 3.16TAVFEE289 pKa = 4.85GSFASYY295 pKa = 9.63TISSSGGVLTISSSASGSDD314 pKa = 3.09RR315 pKa = 11.84ISEE318 pKa = 4.07VEE320 pKa = 3.85RR321 pKa = 11.84FQFADD326 pKa = 3.24STRR329 pKa = 11.84TLDD332 pKa = 3.89QLSADD337 pKa = 4.23SDD339 pKa = 3.79TTAPQLVSLNPLDD352 pKa = 5.18DD353 pKa = 4.47GSNVAVGANLVLSFDD368 pKa = 4.08EE369 pKa = 5.53AIKK372 pKa = 10.75AGNGTISIYY381 pKa = 11.05NSDD384 pKa = 3.31GSLARR389 pKa = 11.84SINAVDD395 pKa = 3.44ATQVRR400 pKa = 11.84INGSTVIVDD409 pKa = 4.03PATDD413 pKa = 4.16LLASRR418 pKa = 11.84GYY420 pKa = 10.92YY421 pKa = 8.08ITLSAGALTDD431 pKa = 4.49LADD434 pKa = 3.84NPFAGISGATRR445 pKa = 11.84WNFNTGSTDD454 pKa = 2.79SSAPQIVALTPADD467 pKa = 3.64DD468 pKa = 4.11SGNVARR474 pKa = 11.84NANLVIVFNEE484 pKa = 4.13SVRR487 pKa = 11.84TGSGNLNIRR496 pKa = 11.84DD497 pKa = 3.77ALGEE501 pKa = 4.04LRR503 pKa = 11.84TIAVTDD509 pKa = 4.29ASQVTIDD516 pKa = 4.07GSTVTINPTADD527 pKa = 3.42LAAGASYY534 pKa = 9.78TITVDD539 pKa = 3.0AGAFHH544 pKa = 7.54DD545 pKa = 5.04LAGNPHH551 pKa = 6.49GGIMTTSAWNFSTSATTVTDD571 pKa = 4.35DD572 pKa = 3.74YY573 pKa = 10.92PYY575 pKa = 10.28STDD578 pKa = 3.21TPGLVVVNGASASGTIEE595 pKa = 3.99VPNDD599 pKa = 3.01QDD601 pKa = 3.91LLRR604 pKa = 11.84VEE606 pKa = 4.99LLAGVNYY613 pKa = 9.63TFDD616 pKa = 3.94LQRR619 pKa = 11.84KK620 pKa = 8.82AGGLADD626 pKa = 3.97PFLVLFSPTITQVAFDD642 pKa = 5.02DD643 pKa = 4.67DD644 pKa = 4.5SGGSGNARR652 pKa = 11.84ISYY655 pKa = 7.73TALTTGSYY663 pKa = 10.17YY664 pKa = 10.77LAVVDD669 pKa = 4.23YY670 pKa = 11.17GIGTGGYY677 pKa = 6.43TLRR680 pKa = 11.84ATTADD685 pKa = 3.39SVAPTLVNRR694 pKa = 11.84TPADD698 pKa = 3.36DD699 pKa = 3.83TQAVSVSADD708 pKa = 3.37LVLDD712 pKa = 4.26FSEE715 pKa = 4.7TVLAGSGSLRR725 pKa = 11.84IYY727 pKa = 10.48NSNGSLVRR735 pKa = 11.84EE736 pKa = 4.15IHH738 pKa = 6.49ASDD741 pKa = 3.44SAGVRR746 pKa = 11.84IAGNRR751 pKa = 11.84VTLNPGEE758 pKa = 4.17NLPAGTSFYY767 pKa = 11.8VNIDD771 pKa = 3.05ANAFRR776 pKa = 11.84DD777 pKa = 3.88ASGNTYY783 pKa = 10.9AGLFDD788 pKa = 3.52TSSWNFSTAAVTSTDD803 pKa = 4.64DD804 pKa = 3.71YY805 pKa = 11.26PLSLNTTGMVTVNGPALNARR825 pKa = 11.84IDD827 pKa = 3.63SSNDD831 pKa = 2.53GDD833 pKa = 4.29LFKK836 pKa = 11.09VDD838 pKa = 4.41LSTGVTYY845 pKa = 10.71LFDD848 pKa = 4.44MISPLSSAVDD858 pKa = 3.73PFLVLYY864 pKa = 10.34GMQPEE869 pKa = 4.6VNLITYY875 pKa = 10.39DD876 pKa = 3.88DD877 pKa = 4.85DD878 pKa = 5.38GGPLPLDD885 pKa = 3.12SRR887 pKa = 11.84IYY889 pKa = 7.97YY890 pKa = 8.53TPSTSGTYY898 pKa = 10.42YY899 pKa = 10.81LAAYY903 pKa = 9.49DD904 pKa = 3.91YY905 pKa = 11.77AEE907 pKa = 4.18ATGTYY912 pKa = 9.39SISATVPVDD921 pKa = 3.92DD922 pKa = 4.84YY923 pKa = 11.65LGSTATSGRR932 pKa = 11.84LVSGGAVLTGSIGVPSDD949 pKa = 2.66IDD951 pKa = 3.6MFALSTMAGVQYY963 pKa = 10.94SVDD966 pKa = 3.65LRR968 pKa = 11.84STGLTDD974 pKa = 4.24PYY976 pKa = 11.21LVLLDD981 pKa = 4.3ADD983 pKa = 4.19GTAIAYY989 pKa = 10.1DD990 pKa = 4.19DD991 pKa = 5.21DD992 pKa = 4.89SLGSLNAEE1000 pKa = 3.9ITFTAQTSGTMFLAVSDD1017 pKa = 4.21FDD1019 pKa = 4.24VGTGAYY1025 pKa = 9.76SINAFPRR1032 pKa = 11.84VANITGTSGGDD1043 pKa = 3.49KK1044 pKa = 10.71LIGGADD1050 pKa = 3.28SDD1052 pKa = 4.97LIYY1055 pKa = 11.11GLGGNDD1061 pKa = 3.66TLTGGAGNDD1070 pKa = 3.7TLGGGDD1076 pKa = 4.63GIDD1079 pKa = 3.73TINYY1083 pKa = 7.14GGPASRR1089 pKa = 11.84YY1090 pKa = 7.81VLGVPGTGWSVQDD1103 pKa = 3.6TSSSEE1108 pKa = 4.03GRR1110 pKa = 11.84DD1111 pKa = 3.33LLYY1114 pKa = 10.88DD1115 pKa = 3.34FEE1117 pKa = 6.91RR1118 pKa = 11.84IHH1120 pKa = 7.38FSDD1123 pKa = 3.77YY1124 pKa = 11.34NLAVDD1129 pKa = 4.05IDD1131 pKa = 4.02GHH1133 pKa = 6.92AGTTAKK1139 pKa = 10.45ILGAVFGATAVHH1151 pKa = 5.85NRR1153 pKa = 11.84TYY1155 pKa = 11.26VGIALGLLDD1164 pKa = 5.47GGMSDD1169 pKa = 3.73QSLMQLALDD1178 pKa = 3.91VRR1180 pKa = 11.84LGTAASHH1187 pKa = 5.46QAVASLLYY1195 pKa = 10.23TNVIGSAPPPDD1206 pKa = 3.5ALAYY1210 pKa = 7.61FTSLLDD1216 pKa = 3.83NGVLTPSGLGVLAAEE1231 pKa = 4.61TTQNAVNINLVGLYY1245 pKa = 9.89EE1246 pKa = 4.28SGIEE1250 pKa = 4.16FVV1252 pKa = 4.05
MM1 pKa = 7.57TIANLTLAQVLDD13 pKa = 3.8QLNSGRR19 pKa = 11.84SWSGASISYY28 pKa = 9.32SFPNTAPGLYY38 pKa = 9.92SQGEE42 pKa = 4.03AAAFRR47 pKa = 11.84AFDD50 pKa = 3.82ADD52 pKa = 3.54QRR54 pKa = 11.84SMMTLAMVTWDD65 pKa = 5.05DD66 pKa = 4.57LISPNFVQGTAGTTHH81 pKa = 7.43IEE83 pKa = 4.03FGYY86 pKa = 6.12TTSXIGFAHH95 pKa = 7.55AYY97 pKa = 8.33YY98 pKa = 10.17PEE100 pKa = 4.68IFGPRR105 pKa = 11.84YY106 pKa = 8.57AAPYY110 pKa = 8.72YY111 pKa = 10.46SSEE114 pKa = 4.02VMQADD119 pKa = 3.47WTGSDD124 pKa = 2.88GRR126 pKa = 11.84TYY128 pKa = 10.86SAQTPMLNDD137 pKa = 2.99VMAIQAIYY145 pKa = 9.65GASTSTRR152 pKa = 11.84LDD154 pKa = 3.11NTVYY158 pKa = 10.77GFGSNIVDD166 pKa = 3.79STRR169 pKa = 11.84AIYY172 pKa = 10.36DD173 pKa = 3.62FSLNAHH179 pKa = 7.61PILTLYY185 pKa = 10.61DD186 pKa = 3.54AGGLDD191 pKa = 3.97TLNLSGWSTPSRR203 pKa = 11.84IDD205 pKa = 3.02LHH207 pKa = 8.18AGAFSSGNDD216 pKa = 3.21MSNNIAIAYY225 pKa = 7.55NATIEE230 pKa = 4.05NAEE233 pKa = 4.18GGGGNDD239 pKa = 3.69VITGNDD245 pKa = 3.31TANLLRR251 pKa = 11.84GAAGNDD257 pKa = 3.36EE258 pKa = 5.1LYY260 pKa = 11.15GSAGDD265 pKa = 3.75DD266 pKa = 3.52TLIGGTGNDD275 pKa = 4.36DD276 pKa = 3.56LRR278 pKa = 11.84GGAGTDD284 pKa = 3.16TAVFEE289 pKa = 4.85GSFASYY295 pKa = 9.63TISSSGGVLTISSSASGSDD314 pKa = 3.09RR315 pKa = 11.84ISEE318 pKa = 4.07VEE320 pKa = 3.85RR321 pKa = 11.84FQFADD326 pKa = 3.24STRR329 pKa = 11.84TLDD332 pKa = 3.89QLSADD337 pKa = 4.23SDD339 pKa = 3.79TTAPQLVSLNPLDD352 pKa = 5.18DD353 pKa = 4.47GSNVAVGANLVLSFDD368 pKa = 4.08EE369 pKa = 5.53AIKK372 pKa = 10.75AGNGTISIYY381 pKa = 11.05NSDD384 pKa = 3.31GSLARR389 pKa = 11.84SINAVDD395 pKa = 3.44ATQVRR400 pKa = 11.84INGSTVIVDD409 pKa = 4.03PATDD413 pKa = 4.16LLASRR418 pKa = 11.84GYY420 pKa = 10.92YY421 pKa = 8.08ITLSAGALTDD431 pKa = 4.49LADD434 pKa = 3.84NPFAGISGATRR445 pKa = 11.84WNFNTGSTDD454 pKa = 2.79SSAPQIVALTPADD467 pKa = 3.64DD468 pKa = 4.11SGNVARR474 pKa = 11.84NANLVIVFNEE484 pKa = 4.13SVRR487 pKa = 11.84TGSGNLNIRR496 pKa = 11.84DD497 pKa = 3.77ALGEE501 pKa = 4.04LRR503 pKa = 11.84TIAVTDD509 pKa = 4.29ASQVTIDD516 pKa = 4.07GSTVTINPTADD527 pKa = 3.42LAAGASYY534 pKa = 9.78TITVDD539 pKa = 3.0AGAFHH544 pKa = 7.54DD545 pKa = 5.04LAGNPHH551 pKa = 6.49GGIMTTSAWNFSTSATTVTDD571 pKa = 4.35DD572 pKa = 3.74YY573 pKa = 10.92PYY575 pKa = 10.28STDD578 pKa = 3.21TPGLVVVNGASASGTIEE595 pKa = 3.99VPNDD599 pKa = 3.01QDD601 pKa = 3.91LLRR604 pKa = 11.84VEE606 pKa = 4.99LLAGVNYY613 pKa = 9.63TFDD616 pKa = 3.94LQRR619 pKa = 11.84KK620 pKa = 8.82AGGLADD626 pKa = 3.97PFLVLFSPTITQVAFDD642 pKa = 5.02DD643 pKa = 4.67DD644 pKa = 4.5SGGSGNARR652 pKa = 11.84ISYY655 pKa = 7.73TALTTGSYY663 pKa = 10.17YY664 pKa = 10.77LAVVDD669 pKa = 4.23YY670 pKa = 11.17GIGTGGYY677 pKa = 6.43TLRR680 pKa = 11.84ATTADD685 pKa = 3.39SVAPTLVNRR694 pKa = 11.84TPADD698 pKa = 3.36DD699 pKa = 3.83TQAVSVSADD708 pKa = 3.37LVLDD712 pKa = 4.26FSEE715 pKa = 4.7TVLAGSGSLRR725 pKa = 11.84IYY727 pKa = 10.48NSNGSLVRR735 pKa = 11.84EE736 pKa = 4.15IHH738 pKa = 6.49ASDD741 pKa = 3.44SAGVRR746 pKa = 11.84IAGNRR751 pKa = 11.84VTLNPGEE758 pKa = 4.17NLPAGTSFYY767 pKa = 11.8VNIDD771 pKa = 3.05ANAFRR776 pKa = 11.84DD777 pKa = 3.88ASGNTYY783 pKa = 10.9AGLFDD788 pKa = 3.52TSSWNFSTAAVTSTDD803 pKa = 4.64DD804 pKa = 3.71YY805 pKa = 11.26PLSLNTTGMVTVNGPALNARR825 pKa = 11.84IDD827 pKa = 3.63SSNDD831 pKa = 2.53GDD833 pKa = 4.29LFKK836 pKa = 11.09VDD838 pKa = 4.41LSTGVTYY845 pKa = 10.71LFDD848 pKa = 4.44MISPLSSAVDD858 pKa = 3.73PFLVLYY864 pKa = 10.34GMQPEE869 pKa = 4.6VNLITYY875 pKa = 10.39DD876 pKa = 3.88DD877 pKa = 4.85DD878 pKa = 5.38GGPLPLDD885 pKa = 3.12SRR887 pKa = 11.84IYY889 pKa = 7.97YY890 pKa = 8.53TPSTSGTYY898 pKa = 10.42YY899 pKa = 10.81LAAYY903 pKa = 9.49DD904 pKa = 3.91YY905 pKa = 11.77AEE907 pKa = 4.18ATGTYY912 pKa = 9.39SISATVPVDD921 pKa = 3.92DD922 pKa = 4.84YY923 pKa = 11.65LGSTATSGRR932 pKa = 11.84LVSGGAVLTGSIGVPSDD949 pKa = 2.66IDD951 pKa = 3.6MFALSTMAGVQYY963 pKa = 10.94SVDD966 pKa = 3.65LRR968 pKa = 11.84STGLTDD974 pKa = 4.24PYY976 pKa = 11.21LVLLDD981 pKa = 4.3ADD983 pKa = 4.19GTAIAYY989 pKa = 10.1DD990 pKa = 4.19DD991 pKa = 5.21DD992 pKa = 4.89SLGSLNAEE1000 pKa = 3.9ITFTAQTSGTMFLAVSDD1017 pKa = 4.21FDD1019 pKa = 4.24VGTGAYY1025 pKa = 9.76SINAFPRR1032 pKa = 11.84VANITGTSGGDD1043 pKa = 3.49KK1044 pKa = 10.71LIGGADD1050 pKa = 3.28SDD1052 pKa = 4.97LIYY1055 pKa = 11.11GLGGNDD1061 pKa = 3.66TLTGGAGNDD1070 pKa = 3.7TLGGGDD1076 pKa = 4.63GIDD1079 pKa = 3.73TINYY1083 pKa = 7.14GGPASRR1089 pKa = 11.84YY1090 pKa = 7.81VLGVPGTGWSVQDD1103 pKa = 3.6TSSSEE1108 pKa = 4.03GRR1110 pKa = 11.84DD1111 pKa = 3.33LLYY1114 pKa = 10.88DD1115 pKa = 3.34FEE1117 pKa = 6.91RR1118 pKa = 11.84IHH1120 pKa = 7.38FSDD1123 pKa = 3.77YY1124 pKa = 11.34NLAVDD1129 pKa = 4.05IDD1131 pKa = 4.02GHH1133 pKa = 6.92AGTTAKK1139 pKa = 10.45ILGAVFGATAVHH1151 pKa = 5.85NRR1153 pKa = 11.84TYY1155 pKa = 11.26VGIALGLLDD1164 pKa = 5.47GGMSDD1169 pKa = 3.73QSLMQLALDD1178 pKa = 3.91VRR1180 pKa = 11.84LGTAASHH1187 pKa = 5.46QAVASLLYY1195 pKa = 10.23TNVIGSAPPPDD1206 pKa = 3.5ALAYY1210 pKa = 7.61FTSLLDD1216 pKa = 3.83NGVLTPSGLGVLAAEE1231 pKa = 4.61TTQNAVNINLVGLYY1245 pKa = 9.89EE1246 pKa = 4.28SGIEE1250 pKa = 4.16FVV1252 pKa = 4.05
Molecular weight: 129.48 kDa
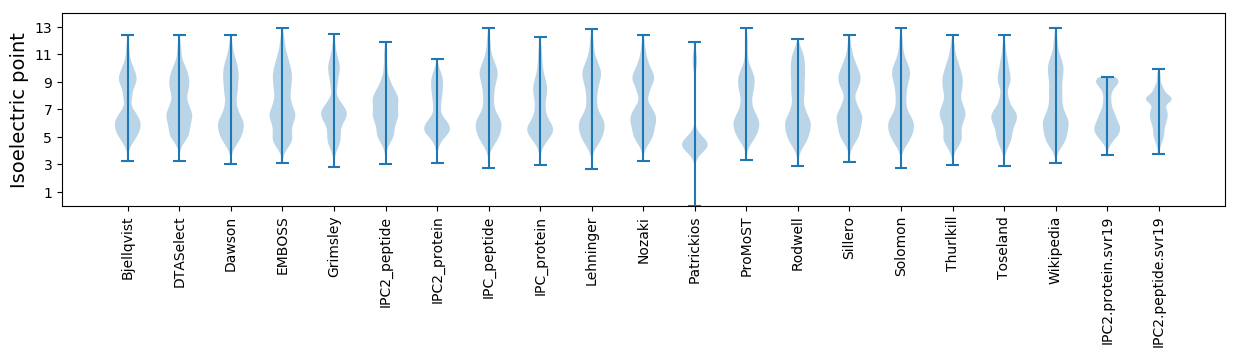
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A080LYM5|A0A080LYM5_9PROT Uncharacterized protein OS=Candidatus Accumulibacter sp. BA-91 OX=1454002 GN=AW09_000759 PE=4 SV=1
MM1 pKa = 7.67LGQLARR7 pKa = 11.84RR8 pKa = 11.84ARR10 pKa = 11.84VDD12 pKa = 3.21WRR14 pKa = 11.84DD15 pKa = 2.75HH16 pKa = 5.82WGFEE20 pKa = 4.16LLRR23 pKa = 11.84LEE25 pKa = 4.41TFVDD29 pKa = 4.11PRR31 pKa = 11.84HH32 pKa = 6.11HH33 pKa = 7.55AGTCYY38 pKa = 10.21LAAGWQLLGVTSRR51 pKa = 11.84RR52 pKa = 11.84GLARR56 pKa = 11.84PSQSYY61 pKa = 10.47HH62 pKa = 4.3STPRR66 pKa = 11.84QVWVKK71 pKa = 9.53PLTSFRR77 pKa = 11.84LLRR80 pKa = 11.84DD81 pKa = 2.99LDD83 pKa = 4.12LAHH86 pKa = 7.3LEE88 pKa = 4.12QAHH91 pKa = 5.77VDD93 pKa = 3.67LVKK96 pKa = 10.75RR97 pKa = 11.84LIRR100 pKa = 11.84GKK102 pKa = 8.74SFRR105 pKa = 11.84RR106 pKa = 11.84YY107 pKa = 9.9LINHH111 pKa = 7.18CDD113 pKa = 3.88PIAIDD118 pKa = 3.95GSLAIPLGGGTPSAPSARR136 pKa = 11.84TKK138 pKa = 10.48PVTPSICSCSTDD150 pKa = 3.16STPTGRR156 pKa = 4.5
MM1 pKa = 7.67LGQLARR7 pKa = 11.84RR8 pKa = 11.84ARR10 pKa = 11.84VDD12 pKa = 3.21WRR14 pKa = 11.84DD15 pKa = 2.75HH16 pKa = 5.82WGFEE20 pKa = 4.16LLRR23 pKa = 11.84LEE25 pKa = 4.41TFVDD29 pKa = 4.11PRR31 pKa = 11.84HH32 pKa = 6.11HH33 pKa = 7.55AGTCYY38 pKa = 10.21LAAGWQLLGVTSRR51 pKa = 11.84RR52 pKa = 11.84GLARR56 pKa = 11.84PSQSYY61 pKa = 10.47HH62 pKa = 4.3STPRR66 pKa = 11.84QVWVKK71 pKa = 9.53PLTSFRR77 pKa = 11.84LLRR80 pKa = 11.84DD81 pKa = 2.99LDD83 pKa = 4.12LAHH86 pKa = 7.3LEE88 pKa = 4.12QAHH91 pKa = 5.77VDD93 pKa = 3.67LVKK96 pKa = 10.75RR97 pKa = 11.84LIRR100 pKa = 11.84GKK102 pKa = 8.74SFRR105 pKa = 11.84RR106 pKa = 11.84YY107 pKa = 9.9LINHH111 pKa = 7.18CDD113 pKa = 3.88PIAIDD118 pKa = 3.95GSLAIPLGGGTPSAPSARR136 pKa = 11.84TKK138 pKa = 10.48PVTPSICSCSTDD150 pKa = 3.16STPTGRR156 pKa = 4.5
Molecular weight: 17.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1187751 |
29 |
1810 |
267.0 |
29.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.926 ± 0.048 | 1.19 ± 0.013 |
5.436 ± 0.034 | 5.677 ± 0.038 |
3.604 ± 0.025 | 7.991 ± 0.041 |
2.435 ± 0.023 | 4.737 ± 0.027 |
2.972 ± 0.032 | 11.201 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.144 ± 0.018 | 2.573 ± 0.028 |
5.062 ± 0.036 | 4.163 ± 0.025 |
7.778 ± 0.046 | 5.588 ± 0.043 |
4.769 ± 0.035 | 7.139 ± 0.036 |
1.349 ± 0.018 | 2.153 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
