
Microbulbifer thermotolerans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Microbulbiferaceae; Microbulbifer
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
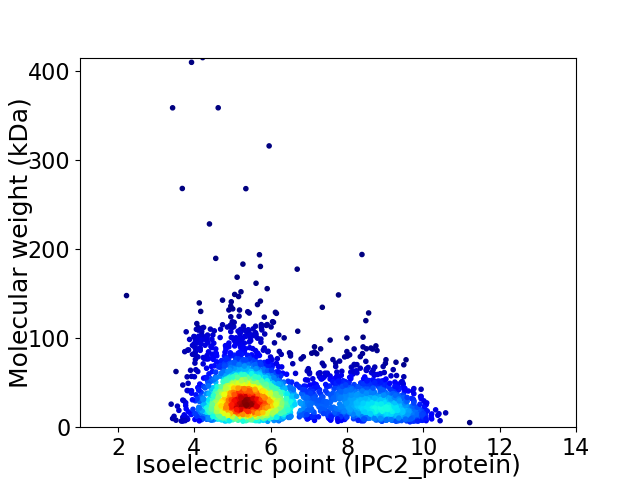
Virtual 2D-PAGE plot for 3126 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
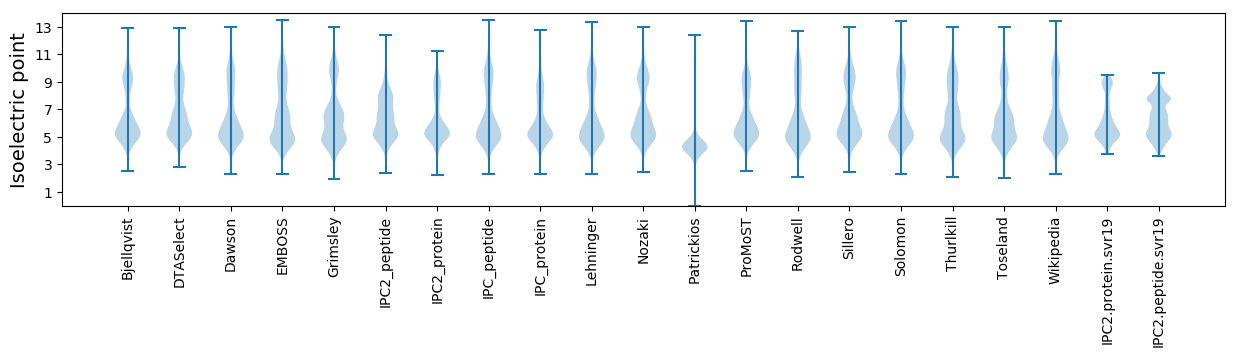
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143HN24|A0A143HN24_9GAMM Cysteine desulfurase IscS OS=Microbulbifer thermotolerans OX=252514 GN=iscS PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.45KK3 pKa = 9.21TILRR7 pKa = 11.84RR8 pKa = 11.84AALAAAISTTSSGAMAAGFYY28 pKa = 10.85LNEE31 pKa = 4.14TSISGLGRR39 pKa = 11.84AFAAEE44 pKa = 3.96NTVGDD49 pKa = 3.85NAAILARR56 pKa = 11.84NPAGSALFDD65 pKa = 4.37TITLSGGVIYY75 pKa = 10.1VNPEE79 pKa = 2.82IDD81 pKa = 3.5AEE83 pKa = 4.38GDD85 pKa = 3.42VTYY88 pKa = 9.99FAAPDD93 pKa = 3.74TPIATLSDD101 pKa = 3.6QKK103 pKa = 10.93ATDD106 pKa = 3.81YY107 pKa = 11.59APTAWVPNAYY117 pKa = 10.16LAVPFNEE124 pKa = 3.67NWSFGLAMYY133 pKa = 10.66SNYY136 pKa = 10.58GLEE139 pKa = 4.17TDD141 pKa = 5.88FDD143 pKa = 5.26SDD145 pKa = 3.81WPVTTIADD153 pKa = 3.72KK154 pKa = 11.37TEE156 pKa = 4.0LLTVNIAPSVAYY168 pKa = 10.04DD169 pKa = 3.62FNDD172 pKa = 3.12QFSIGFAVNFLYY184 pKa = 10.84ADD186 pKa = 3.67ATLKK190 pKa = 10.38TMVPTNFGLNPTVAGADD207 pKa = 3.01ILSYY211 pKa = 11.44DD212 pKa = 5.07DD213 pKa = 4.63ADD215 pKa = 3.6DD216 pKa = 4.5WEE218 pKa = 4.6VSWTIGALWNITDD231 pKa = 3.65RR232 pKa = 11.84TRR234 pKa = 11.84FGISYY239 pKa = 9.59HH240 pKa = 6.96AEE242 pKa = 3.88LDD244 pKa = 3.74PEE246 pKa = 4.53LEE248 pKa = 4.28GDD250 pKa = 3.97VNSDD254 pKa = 4.04LVPLPLSPFNDD265 pKa = 3.16AKK267 pKa = 11.33GKK269 pKa = 8.37VTLDD273 pKa = 3.59LPDD276 pKa = 3.69TLEE279 pKa = 4.16LGLYY283 pKa = 10.11HH284 pKa = 7.49RR285 pKa = 11.84FNEE288 pKa = 4.04NWGLALGALWTDD300 pKa = 2.5WDD302 pKa = 3.82DD303 pKa = 4.51FEE305 pKa = 5.98RR306 pKa = 11.84LEE308 pKa = 4.23AFIPAGGEE316 pKa = 4.03GFNPLLFKK324 pKa = 10.98EE325 pKa = 4.88EE326 pKa = 4.08NFQSGWRR333 pKa = 11.84YY334 pKa = 10.32SIAGEE339 pKa = 4.16YY340 pKa = 9.23YY341 pKa = 8.21PCEE344 pKa = 4.03EE345 pKa = 3.77VTLRR349 pKa = 11.84IGYY352 pKa = 10.23AYY354 pKa = 10.33DD355 pKa = 3.18EE356 pKa = 4.69GAARR360 pKa = 11.84DD361 pKa = 3.91GFNTGDD367 pKa = 3.59AAANAYY373 pKa = 8.37GLPITWRR380 pKa = 11.84TLSIPDD386 pKa = 3.6TDD388 pKa = 4.47RR389 pKa = 11.84NWVTIGGTYY398 pKa = 10.36KK399 pKa = 10.66FNNQLSIDD407 pKa = 3.88GGLAYY412 pKa = 10.61LWGDD416 pKa = 3.7DD417 pKa = 3.48EE418 pKa = 5.03TIQEE422 pKa = 4.57FTTVPAPTYY431 pKa = 10.64FDD433 pKa = 4.04GGTTNIEE440 pKa = 3.36AWLAGVSLNYY450 pKa = 10.71AFF452 pKa = 5.5
MM1 pKa = 7.71KK2 pKa = 9.45KK3 pKa = 9.21TILRR7 pKa = 11.84RR8 pKa = 11.84AALAAAISTTSSGAMAAGFYY28 pKa = 10.85LNEE31 pKa = 4.14TSISGLGRR39 pKa = 11.84AFAAEE44 pKa = 3.96NTVGDD49 pKa = 3.85NAAILARR56 pKa = 11.84NPAGSALFDD65 pKa = 4.37TITLSGGVIYY75 pKa = 10.1VNPEE79 pKa = 2.82IDD81 pKa = 3.5AEE83 pKa = 4.38GDD85 pKa = 3.42VTYY88 pKa = 9.99FAAPDD93 pKa = 3.74TPIATLSDD101 pKa = 3.6QKK103 pKa = 10.93ATDD106 pKa = 3.81YY107 pKa = 11.59APTAWVPNAYY117 pKa = 10.16LAVPFNEE124 pKa = 3.67NWSFGLAMYY133 pKa = 10.66SNYY136 pKa = 10.58GLEE139 pKa = 4.17TDD141 pKa = 5.88FDD143 pKa = 5.26SDD145 pKa = 3.81WPVTTIADD153 pKa = 3.72KK154 pKa = 11.37TEE156 pKa = 4.0LLTVNIAPSVAYY168 pKa = 10.04DD169 pKa = 3.62FNDD172 pKa = 3.12QFSIGFAVNFLYY184 pKa = 10.84ADD186 pKa = 3.67ATLKK190 pKa = 10.38TMVPTNFGLNPTVAGADD207 pKa = 3.01ILSYY211 pKa = 11.44DD212 pKa = 5.07DD213 pKa = 4.63ADD215 pKa = 3.6DD216 pKa = 4.5WEE218 pKa = 4.6VSWTIGALWNITDD231 pKa = 3.65RR232 pKa = 11.84TRR234 pKa = 11.84FGISYY239 pKa = 9.59HH240 pKa = 6.96AEE242 pKa = 3.88LDD244 pKa = 3.74PEE246 pKa = 4.53LEE248 pKa = 4.28GDD250 pKa = 3.97VNSDD254 pKa = 4.04LVPLPLSPFNDD265 pKa = 3.16AKK267 pKa = 11.33GKK269 pKa = 8.37VTLDD273 pKa = 3.59LPDD276 pKa = 3.69TLEE279 pKa = 4.16LGLYY283 pKa = 10.11HH284 pKa = 7.49RR285 pKa = 11.84FNEE288 pKa = 4.04NWGLALGALWTDD300 pKa = 2.5WDD302 pKa = 3.82DD303 pKa = 4.51FEE305 pKa = 5.98RR306 pKa = 11.84LEE308 pKa = 4.23AFIPAGGEE316 pKa = 4.03GFNPLLFKK324 pKa = 10.98EE325 pKa = 4.88EE326 pKa = 4.08NFQSGWRR333 pKa = 11.84YY334 pKa = 10.32SIAGEE339 pKa = 4.16YY340 pKa = 9.23YY341 pKa = 8.21PCEE344 pKa = 4.03EE345 pKa = 3.77VTLRR349 pKa = 11.84IGYY352 pKa = 10.23AYY354 pKa = 10.33DD355 pKa = 3.18EE356 pKa = 4.69GAARR360 pKa = 11.84DD361 pKa = 3.91GFNTGDD367 pKa = 3.59AAANAYY373 pKa = 8.37GLPITWRR380 pKa = 11.84TLSIPDD386 pKa = 3.6TDD388 pKa = 4.47RR389 pKa = 11.84NWVTIGGTYY398 pKa = 10.36KK399 pKa = 10.66FNNQLSIDD407 pKa = 3.88GGLAYY412 pKa = 10.61LWGDD416 pKa = 3.7DD417 pKa = 3.48EE418 pKa = 5.03TIQEE422 pKa = 4.57FTTVPAPTYY431 pKa = 10.64FDD433 pKa = 4.04GGTTNIEE440 pKa = 3.36AWLAGVSLNYY450 pKa = 10.71AFF452 pKa = 5.5
Molecular weight: 49.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143HR25|A0A143HR25_9GAMM Uncharacterized protein OS=Microbulbifer thermotolerans OX=252514 GN=A3224_15085 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1082839 |
44 |
3937 |
346.4 |
38.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.265 ± 0.052 | 1.047 ± 0.015 |
5.707 ± 0.051 | 6.661 ± 0.038 |
3.715 ± 0.023 | 7.941 ± 0.048 |
2.099 ± 0.023 | 5.105 ± 0.035 |
3.75 ± 0.038 | 10.745 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.163 ± 0.023 | 3.39 ± 0.034 |
4.647 ± 0.031 | 4.107 ± 0.031 |
6.606 ± 0.046 | 5.929 ± 0.039 |
4.882 ± 0.033 | 6.856 ± 0.038 |
1.434 ± 0.019 | 2.951 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
