
Pontimonas salivibrio
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Pontimonas
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
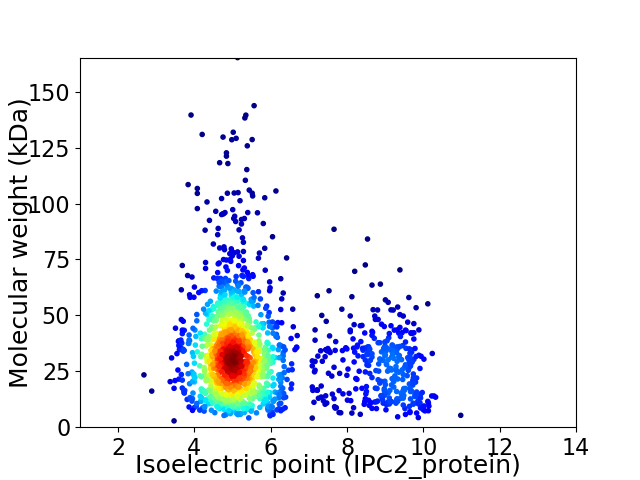
Virtual 2D-PAGE plot for 1693 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L2BQ19|A0A2L2BQ19_9MICO Cation-translocating P-type ATPase MgtA OS=Pontimonas salivibrio OX=1159327 GN=C3B54_11721 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.39KK3 pKa = 9.72ILSLVAISALALTGITALEE22 pKa = 4.15QEE24 pKa = 4.86AGAAADD30 pKa = 3.94PASLSLTCTSNTTLVSSLSDD50 pKa = 3.64EE51 pKa = 4.82FPAGAPYY58 pKa = 10.3VISALIGDD66 pKa = 5.36DD67 pKa = 3.35IDD69 pKa = 3.92VTASGTSTDD78 pKa = 4.49DD79 pKa = 3.71DD80 pKa = 4.4CDD82 pKa = 3.6FANVSGNAALTITGGGTVEE101 pKa = 4.97GGTSTAANEE110 pKa = 3.89LDD112 pKa = 3.92VANSGAFTITPVTGDD127 pKa = 3.3AVTVYY132 pKa = 10.69VDD134 pKa = 3.31ACSLSGSGITSDD146 pKa = 2.96PWLVGTAADD155 pKa = 4.32FKK157 pKa = 11.33SVGAEE162 pKa = 3.63SSTAGEE168 pKa = 4.31NSCSLAGHH176 pKa = 6.12YY177 pKa = 9.95LQTASLTGVDD187 pKa = 3.72TSSVGDD193 pKa = 3.73SDD195 pKa = 4.36WIVDD199 pKa = 3.87GVFTGTYY206 pKa = 10.4DD207 pKa = 3.52GDD209 pKa = 4.06HH210 pKa = 6.48YY211 pKa = 11.18SISYY215 pKa = 9.96FSGGGSATFQGRR227 pKa = 11.84SPVFDD232 pKa = 3.69EE233 pKa = 4.96LGSGGVIKK241 pKa = 10.66RR242 pKa = 11.84LNLTGHH248 pKa = 6.83IKK250 pKa = 10.77GDD252 pKa = 3.6STRR255 pKa = 11.84NASLVEE261 pKa = 3.92YY262 pKa = 10.18LVGGTISEE270 pKa = 4.34VRR272 pKa = 11.84SSVWLEE278 pKa = 4.04VEE280 pKa = 5.02DD281 pKa = 4.28SNALIGGLAAVTGSSSVVSDD301 pKa = 3.63ASSRR305 pKa = 11.84IQYY308 pKa = 9.88SKK310 pKa = 10.84YY311 pKa = 9.73SGRR314 pKa = 11.84IDD316 pKa = 3.23WVDD319 pKa = 3.68DD320 pKa = 3.51GTGDD324 pKa = 4.29VEE326 pKa = 4.63GPTIGGLVGMVVGTGTTEE344 pKa = 4.01LRR346 pKa = 11.84DD347 pKa = 3.96SYY349 pKa = 11.64SLASISYY356 pKa = 10.54DD357 pKa = 3.52SGDD360 pKa = 3.69LTGTDD365 pKa = 4.25PDD367 pKa = 3.85TAIYY371 pKa = 10.52AGGLVGSDD379 pKa = 3.76GEE381 pKa = 4.27TDD383 pKa = 3.42LAAFNTDD390 pKa = 3.17DD391 pKa = 4.07GDD393 pKa = 3.93RR394 pKa = 11.84TFSASEE400 pKa = 3.69VRR402 pKa = 11.84IVRR405 pKa = 11.84SYY407 pKa = 10.91FAGSFSNLCEE417 pKa = 4.69GSAADD422 pKa = 4.9CNTDD426 pKa = 3.3SPSHH430 pKa = 5.96VFTGGLFGVSEE441 pKa = 4.93DD442 pKa = 5.43LDD444 pKa = 4.3DD445 pKa = 6.25ADD447 pKa = 5.3DD448 pKa = 4.75LLVSAFWLSSSASNAVGEE466 pKa = 4.34IVDD469 pKa = 5.33AGTQPLDD476 pKa = 3.44YY477 pKa = 10.17TAATPALPEE486 pKa = 4.25APGLSATLLKK496 pKa = 9.91TVSTFQSEE504 pKa = 4.48EE505 pKa = 4.05GSTTGSPSGDD515 pKa = 3.3ADD517 pKa = 4.18LLVASSTGTLAEE529 pKa = 3.74QDD531 pKa = 3.84YY532 pKa = 10.55RR533 pKa = 11.84WAIEE537 pKa = 3.98SGNIQTFVPSDD548 pKa = 3.58YY549 pKa = 10.62TSEE552 pKa = 4.26ANYY555 pKa = 8.62LTRR558 pKa = 11.84QLFSDD563 pKa = 4.0TSVSQSYY570 pKa = 9.7RR571 pKa = 11.84RR572 pKa = 11.84EE573 pKa = 3.95GAGDD577 pKa = 3.73LTVHH581 pKa = 6.65GGDD584 pKa = 4.26DD585 pKa = 3.9PEE587 pKa = 4.75SVTGYY592 pKa = 6.83PTLGRR597 pKa = 11.84VWEE600 pKa = 4.37VCSNSNSGYY609 pKa = 9.78PFLVWEE615 pKa = 4.8EE616 pKa = 4.45LDD618 pKa = 3.9CSATGGGSDD627 pKa = 5.73DD628 pKa = 5.64DD629 pKa = 6.16DD630 pKa = 6.78DD631 pKa = 6.42DD632 pKa = 4.81EE633 pKa = 6.19SSEE636 pKa = 4.16SSAAALGLTEE646 pKa = 4.6AEE648 pKa = 4.12YY649 pKa = 10.95LAFLASGLTLEE660 pKa = 4.28QFKK663 pKa = 10.47AARR666 pKa = 11.84LAATGPSNEE675 pKa = 3.7TMALGFYY682 pKa = 9.77STVLLVGMGLVLLWGYY698 pKa = 10.11RR699 pKa = 11.84RR700 pKa = 11.84QRR702 pKa = 11.84AEE704 pKa = 3.66SSS706 pKa = 3.12
MM1 pKa = 7.47KK2 pKa = 10.39KK3 pKa = 9.72ILSLVAISALALTGITALEE22 pKa = 4.15QEE24 pKa = 4.86AGAAADD30 pKa = 3.94PASLSLTCTSNTTLVSSLSDD50 pKa = 3.64EE51 pKa = 4.82FPAGAPYY58 pKa = 10.3VISALIGDD66 pKa = 5.36DD67 pKa = 3.35IDD69 pKa = 3.92VTASGTSTDD78 pKa = 4.49DD79 pKa = 3.71DD80 pKa = 4.4CDD82 pKa = 3.6FANVSGNAALTITGGGTVEE101 pKa = 4.97GGTSTAANEE110 pKa = 3.89LDD112 pKa = 3.92VANSGAFTITPVTGDD127 pKa = 3.3AVTVYY132 pKa = 10.69VDD134 pKa = 3.31ACSLSGSGITSDD146 pKa = 2.96PWLVGTAADD155 pKa = 4.32FKK157 pKa = 11.33SVGAEE162 pKa = 3.63SSTAGEE168 pKa = 4.31NSCSLAGHH176 pKa = 6.12YY177 pKa = 9.95LQTASLTGVDD187 pKa = 3.72TSSVGDD193 pKa = 3.73SDD195 pKa = 4.36WIVDD199 pKa = 3.87GVFTGTYY206 pKa = 10.4DD207 pKa = 3.52GDD209 pKa = 4.06HH210 pKa = 6.48YY211 pKa = 11.18SISYY215 pKa = 9.96FSGGGSATFQGRR227 pKa = 11.84SPVFDD232 pKa = 3.69EE233 pKa = 4.96LGSGGVIKK241 pKa = 10.66RR242 pKa = 11.84LNLTGHH248 pKa = 6.83IKK250 pKa = 10.77GDD252 pKa = 3.6STRR255 pKa = 11.84NASLVEE261 pKa = 3.92YY262 pKa = 10.18LVGGTISEE270 pKa = 4.34VRR272 pKa = 11.84SSVWLEE278 pKa = 4.04VEE280 pKa = 5.02DD281 pKa = 4.28SNALIGGLAAVTGSSSVVSDD301 pKa = 3.63ASSRR305 pKa = 11.84IQYY308 pKa = 9.88SKK310 pKa = 10.84YY311 pKa = 9.73SGRR314 pKa = 11.84IDD316 pKa = 3.23WVDD319 pKa = 3.68DD320 pKa = 3.51GTGDD324 pKa = 4.29VEE326 pKa = 4.63GPTIGGLVGMVVGTGTTEE344 pKa = 4.01LRR346 pKa = 11.84DD347 pKa = 3.96SYY349 pKa = 11.64SLASISYY356 pKa = 10.54DD357 pKa = 3.52SGDD360 pKa = 3.69LTGTDD365 pKa = 4.25PDD367 pKa = 3.85TAIYY371 pKa = 10.52AGGLVGSDD379 pKa = 3.76GEE381 pKa = 4.27TDD383 pKa = 3.42LAAFNTDD390 pKa = 3.17DD391 pKa = 4.07GDD393 pKa = 3.93RR394 pKa = 11.84TFSASEE400 pKa = 3.69VRR402 pKa = 11.84IVRR405 pKa = 11.84SYY407 pKa = 10.91FAGSFSNLCEE417 pKa = 4.69GSAADD422 pKa = 4.9CNTDD426 pKa = 3.3SPSHH430 pKa = 5.96VFTGGLFGVSEE441 pKa = 4.93DD442 pKa = 5.43LDD444 pKa = 4.3DD445 pKa = 6.25ADD447 pKa = 5.3DD448 pKa = 4.75LLVSAFWLSSSASNAVGEE466 pKa = 4.34IVDD469 pKa = 5.33AGTQPLDD476 pKa = 3.44YY477 pKa = 10.17TAATPALPEE486 pKa = 4.25APGLSATLLKK496 pKa = 9.91TVSTFQSEE504 pKa = 4.48EE505 pKa = 4.05GSTTGSPSGDD515 pKa = 3.3ADD517 pKa = 4.18LLVASSTGTLAEE529 pKa = 3.74QDD531 pKa = 3.84YY532 pKa = 10.55RR533 pKa = 11.84WAIEE537 pKa = 3.98SGNIQTFVPSDD548 pKa = 3.58YY549 pKa = 10.62TSEE552 pKa = 4.26ANYY555 pKa = 8.62LTRR558 pKa = 11.84QLFSDD563 pKa = 4.0TSVSQSYY570 pKa = 9.7RR571 pKa = 11.84RR572 pKa = 11.84EE573 pKa = 3.95GAGDD577 pKa = 3.73LTVHH581 pKa = 6.65GGDD584 pKa = 4.26DD585 pKa = 3.9PEE587 pKa = 4.75SVTGYY592 pKa = 6.83PTLGRR597 pKa = 11.84VWEE600 pKa = 4.37VCSNSNSGYY609 pKa = 9.78PFLVWEE615 pKa = 4.8EE616 pKa = 4.45LDD618 pKa = 3.9CSATGGGSDD627 pKa = 5.73DD628 pKa = 5.64DD629 pKa = 6.16DD630 pKa = 6.78DD631 pKa = 6.42DD632 pKa = 4.81EE633 pKa = 6.19SSEE636 pKa = 4.16SSAAALGLTEE646 pKa = 4.6AEE648 pKa = 4.12YY649 pKa = 10.95LAFLASGLTLEE660 pKa = 4.28QFKK663 pKa = 10.47AARR666 pKa = 11.84LAATGPSNEE675 pKa = 3.7TMALGFYY682 pKa = 9.77STVLLVGMGLVLLWGYY698 pKa = 10.11RR699 pKa = 11.84RR700 pKa = 11.84QRR702 pKa = 11.84AEE704 pKa = 3.66SSS706 pKa = 3.12
Molecular weight: 72.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L2BN97|A0A2L2BN97_9MICO Foldase YidC OS=Pontimonas salivibrio OX=1159327 GN=C3B54_11139 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILTARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILTARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
538662 |
25 |
1521 |
318.2 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.106 ± 0.071 | 0.487 ± 0.014 |
5.987 ± 0.055 | 6.279 ± 0.05 |
3.31 ± 0.036 | 8.693 ± 0.057 |
2.303 ± 0.033 | 4.933 ± 0.04 |
2.653 ± 0.046 | 10.166 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.019 ± 0.022 | 2.295 ± 0.03 |
5.203 ± 0.04 | 3.464 ± 0.035 |
6.285 ± 0.054 | 6.517 ± 0.046 |
5.865 ± 0.047 | 8.948 ± 0.056 |
1.471 ± 0.028 | 2.015 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
