
Weissella ceti
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Weissella
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
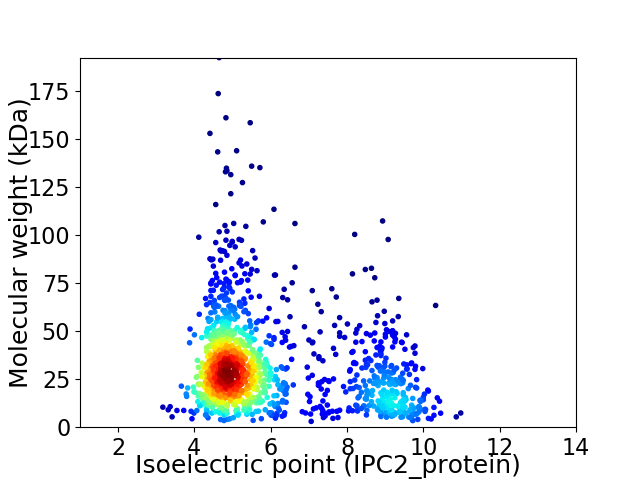
Virtual 2D-PAGE plot for 1338 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
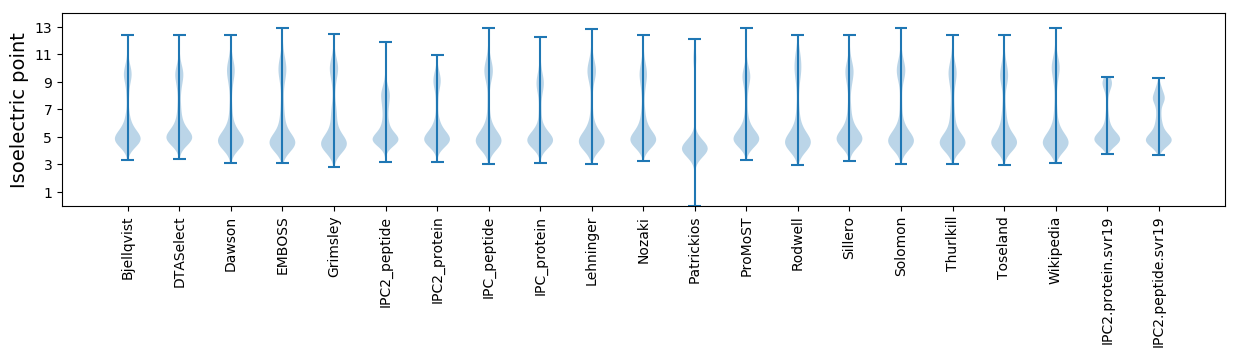
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075TZF1|A0A075TZF1_9LACO Addiction module antitoxin RelB/DinJ family OS=Weissella ceti OX=759620 GN=WS74_0743 PE=3 SV=1
MM1 pKa = 7.61ASFGEE6 pKa = 4.19RR7 pKa = 11.84MLNEE11 pKa = 4.04LALGQVDD18 pKa = 3.81EE19 pKa = 4.83AKK21 pKa = 10.62KK22 pKa = 10.8SFASALRR29 pKa = 11.84HH30 pKa = 6.77DD31 pKa = 5.55DD32 pKa = 4.03DD33 pKa = 3.97DD34 pKa = 4.66TIYY37 pKa = 11.32SLAEE41 pKa = 3.53EE42 pKa = 5.18LYY44 pKa = 10.96ALGMSNQAKK53 pKa = 9.72RR54 pKa = 11.84AYY56 pKa = 9.08EE57 pKa = 3.99KK58 pKa = 10.94LLEE61 pKa = 4.57RR62 pKa = 11.84YY63 pKa = 9.11PDD65 pKa = 3.54EE66 pKa = 4.75DD67 pKa = 3.47QLRR70 pKa = 11.84TALADD75 pKa = 3.37IAIDD79 pKa = 4.73EE80 pKa = 5.33DD81 pKa = 3.98DD82 pKa = 3.62TDD84 pKa = 3.73AAMGYY89 pKa = 10.0LADD92 pKa = 5.07IKK94 pKa = 9.69PTSNAYY100 pKa = 10.36LEE102 pKa = 4.43SLLVLADD109 pKa = 4.59LYY111 pKa = 11.1QSEE114 pKa = 4.37GLYY117 pKa = 10.54EE118 pKa = 3.99PAEE121 pKa = 4.28AKK123 pKa = 10.62LLEE126 pKa = 4.2AHH128 pKa = 6.41NLAPEE133 pKa = 3.89EE134 pKa = 3.97AVIGFALAEE143 pKa = 4.26YY144 pKa = 9.84YY145 pKa = 10.11FASAKK150 pKa = 8.85YY151 pKa = 9.72QEE153 pKa = 5.11AIPYY157 pKa = 9.28YY158 pKa = 10.39RR159 pKa = 11.84EE160 pKa = 3.78LLKK163 pKa = 11.24AGDD166 pKa = 4.28RR167 pKa = 11.84YY168 pKa = 10.73FSGTDD173 pKa = 3.02VASRR177 pKa = 11.84IGVAYY182 pKa = 10.28ALVGDD187 pKa = 4.06VTNALAYY194 pKa = 10.49LEE196 pKa = 4.28QIKK199 pKa = 7.65TTEE202 pKa = 3.96LTPDD206 pKa = 2.88VRR208 pKa = 11.84FQLGMLYY215 pKa = 10.44AADD218 pKa = 4.2EE219 pKa = 4.33EE220 pKa = 4.84THH222 pKa = 6.12QKK224 pKa = 10.91AIDD227 pKa = 3.65TFEE230 pKa = 3.96EE231 pKa = 4.21LMEE234 pKa = 5.96IDD236 pKa = 4.01ASYY239 pKa = 9.04ATLYY243 pKa = 10.52EE244 pKa = 4.07PLGNLYY250 pKa = 10.17EE251 pKa = 4.52HH252 pKa = 7.03AQQDD256 pKa = 3.58EE257 pKa = 4.49DD258 pKa = 4.73ALRR261 pKa = 11.84TYY263 pKa = 10.47QAGLAVDD270 pKa = 3.5QFNTKK275 pKa = 9.83LVEE278 pKa = 3.89RR279 pKa = 11.84AAILSEE285 pKa = 3.87RR286 pKa = 11.84LGSSEE291 pKa = 4.76QAEE294 pKa = 4.11QLYY297 pKa = 10.47KK298 pKa = 10.8EE299 pKa = 4.43GLRR302 pKa = 11.84NNPGDD307 pKa = 3.61PTLVLDD313 pKa = 3.88YY314 pKa = 11.25SDD316 pKa = 3.78WLVLQNKK323 pKa = 8.26YY324 pKa = 10.98AEE326 pKa = 4.85NIALLNDD333 pKa = 3.44YY334 pKa = 10.58LADD337 pKa = 4.64DD338 pKa = 4.69EE339 pKa = 6.19ADD341 pKa = 3.42IDD343 pKa = 3.94PVIYY347 pKa = 10.48RR348 pKa = 11.84NLAQSYY354 pKa = 7.64TALEE358 pKa = 4.7DD359 pKa = 3.67YY360 pKa = 11.3EE361 pKa = 4.61MATQYY366 pKa = 9.18WQAAVPLFIDD376 pKa = 3.81DD377 pKa = 4.52ANFLRR382 pKa = 11.84EE383 pKa = 3.6AFFYY387 pKa = 10.66FRR389 pKa = 11.84EE390 pKa = 4.23NGNQEE395 pKa = 4.1LALEE399 pKa = 4.19TLTRR403 pKa = 11.84YY404 pKa = 9.94VEE406 pKa = 4.21LVPTDD411 pKa = 4.2LEE413 pKa = 4.18MSEE416 pKa = 4.29TLAALADD423 pKa = 3.52TDD425 pKa = 4.16YY426 pKa = 11.97
MM1 pKa = 7.61ASFGEE6 pKa = 4.19RR7 pKa = 11.84MLNEE11 pKa = 4.04LALGQVDD18 pKa = 3.81EE19 pKa = 4.83AKK21 pKa = 10.62KK22 pKa = 10.8SFASALRR29 pKa = 11.84HH30 pKa = 6.77DD31 pKa = 5.55DD32 pKa = 4.03DD33 pKa = 3.97DD34 pKa = 4.66TIYY37 pKa = 11.32SLAEE41 pKa = 3.53EE42 pKa = 5.18LYY44 pKa = 10.96ALGMSNQAKK53 pKa = 9.72RR54 pKa = 11.84AYY56 pKa = 9.08EE57 pKa = 3.99KK58 pKa = 10.94LLEE61 pKa = 4.57RR62 pKa = 11.84YY63 pKa = 9.11PDD65 pKa = 3.54EE66 pKa = 4.75DD67 pKa = 3.47QLRR70 pKa = 11.84TALADD75 pKa = 3.37IAIDD79 pKa = 4.73EE80 pKa = 5.33DD81 pKa = 3.98DD82 pKa = 3.62TDD84 pKa = 3.73AAMGYY89 pKa = 10.0LADD92 pKa = 5.07IKK94 pKa = 9.69PTSNAYY100 pKa = 10.36LEE102 pKa = 4.43SLLVLADD109 pKa = 4.59LYY111 pKa = 11.1QSEE114 pKa = 4.37GLYY117 pKa = 10.54EE118 pKa = 3.99PAEE121 pKa = 4.28AKK123 pKa = 10.62LLEE126 pKa = 4.2AHH128 pKa = 6.41NLAPEE133 pKa = 3.89EE134 pKa = 3.97AVIGFALAEE143 pKa = 4.26YY144 pKa = 9.84YY145 pKa = 10.11FASAKK150 pKa = 8.85YY151 pKa = 9.72QEE153 pKa = 5.11AIPYY157 pKa = 9.28YY158 pKa = 10.39RR159 pKa = 11.84EE160 pKa = 3.78LLKK163 pKa = 11.24AGDD166 pKa = 4.28RR167 pKa = 11.84YY168 pKa = 10.73FSGTDD173 pKa = 3.02VASRR177 pKa = 11.84IGVAYY182 pKa = 10.28ALVGDD187 pKa = 4.06VTNALAYY194 pKa = 10.49LEE196 pKa = 4.28QIKK199 pKa = 7.65TTEE202 pKa = 3.96LTPDD206 pKa = 2.88VRR208 pKa = 11.84FQLGMLYY215 pKa = 10.44AADD218 pKa = 4.2EE219 pKa = 4.33EE220 pKa = 4.84THH222 pKa = 6.12QKK224 pKa = 10.91AIDD227 pKa = 3.65TFEE230 pKa = 3.96EE231 pKa = 4.21LMEE234 pKa = 5.96IDD236 pKa = 4.01ASYY239 pKa = 9.04ATLYY243 pKa = 10.52EE244 pKa = 4.07PLGNLYY250 pKa = 10.17EE251 pKa = 4.52HH252 pKa = 7.03AQQDD256 pKa = 3.58EE257 pKa = 4.49DD258 pKa = 4.73ALRR261 pKa = 11.84TYY263 pKa = 10.47QAGLAVDD270 pKa = 3.5QFNTKK275 pKa = 9.83LVEE278 pKa = 3.89RR279 pKa = 11.84AAILSEE285 pKa = 3.87RR286 pKa = 11.84LGSSEE291 pKa = 4.76QAEE294 pKa = 4.11QLYY297 pKa = 10.47KK298 pKa = 10.8EE299 pKa = 4.43GLRR302 pKa = 11.84NNPGDD307 pKa = 3.61PTLVLDD313 pKa = 3.88YY314 pKa = 11.25SDD316 pKa = 3.78WLVLQNKK323 pKa = 8.26YY324 pKa = 10.98AEE326 pKa = 4.85NIALLNDD333 pKa = 3.44YY334 pKa = 10.58LADD337 pKa = 4.64DD338 pKa = 4.69EE339 pKa = 6.19ADD341 pKa = 3.42IDD343 pKa = 3.94PVIYY347 pKa = 10.48RR348 pKa = 11.84NLAQSYY354 pKa = 7.64TALEE358 pKa = 4.7DD359 pKa = 3.67YY360 pKa = 11.3EE361 pKa = 4.61MATQYY366 pKa = 9.18WQAAVPLFIDD376 pKa = 3.81DD377 pKa = 4.52ANFLRR382 pKa = 11.84EE383 pKa = 3.6AFFYY387 pKa = 10.66FRR389 pKa = 11.84EE390 pKa = 4.23NGNQEE395 pKa = 4.1LALEE399 pKa = 4.19TLTRR403 pKa = 11.84YY404 pKa = 9.94VEE406 pKa = 4.21LVPTDD411 pKa = 4.2LEE413 pKa = 4.18MSEE416 pKa = 4.29TLAALADD423 pKa = 3.52TDD425 pKa = 4.16YY426 pKa = 11.97
Molecular weight: 48.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075TXN0|A0A075TXN0_9LACO Uncharacterized protein OS=Weissella ceti OX=759620 GN=WS74_0148 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.34KK9 pKa = 7.47RR10 pKa = 11.84HH11 pKa = 5.71RR12 pKa = 11.84EE13 pKa = 3.7RR14 pKa = 11.84VHH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
398033 |
29 |
1673 |
297.5 |
33.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.893 ± 0.095 | 0.2 ± 0.012 |
6.208 ± 0.066 | 6.564 ± 0.096 |
4.129 ± 0.049 | 6.926 ± 0.069 |
1.858 ± 0.033 | 6.616 ± 0.061 |
5.487 ± 0.094 | 9.504 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.232 ± 0.038 | 4.463 ± 0.045 |
3.703 ± 0.062 | 4.314 ± 0.062 |
4.219 ± 0.049 | 5.232 ± 0.054 |
6.38 ± 0.054 | 7.759 ± 0.052 |
1.079 ± 0.026 | 3.232 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
